染色质是真核生物细胞核内由核酸和蛋白质组成的复合结构[1],其中核酸包括DNA和少量RNA,蛋白质则包括组蛋白和非组蛋白[2]。
人类基因组序列若线性展开长约2 m,但细胞核只是微米级别,这提示我们DNA势必经过精密且复杂的折叠才能组装进细胞核内。关于染色质的结构,目前主流观点认为:每约147 bp线性DNA首先缠绕八聚核心组蛋白1.75圈组成基本单元核小体[3],核小体再经螺旋盘绕形成30 nm螺线管结构,随后在cohesin和CTCF(CCCTC-binding factor)等蛋白作用下内聚成环(Loops),这样使得线性距离较远的基因座(Chromatin loci)在空间上得以接近,接下来环继续折叠形成拓扑相关结构域(Topologically associated domains,TADs),最终被分配到不同的隔室(Compartment A/B)[4]。
鉴于染色质结构和功能的复杂性,染色质测序涉及多种NGS技术,主要包括以下方面(图 1):
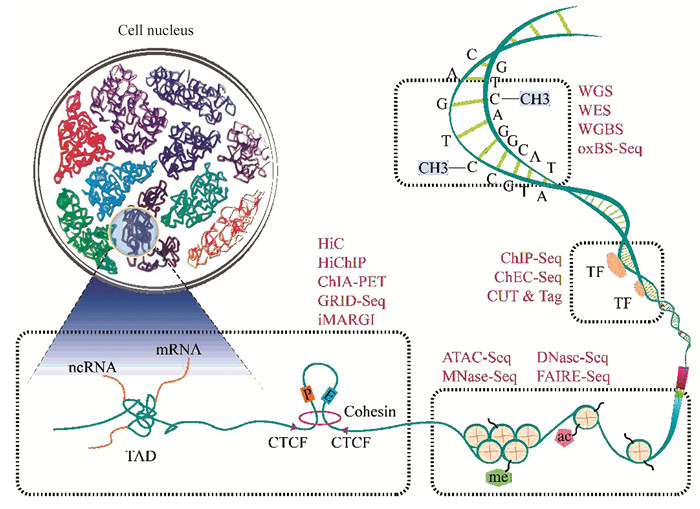
|
图 1 染色质不同层次结构及相应测序技术 Figure 1 Different hierarchical chromatin structures and corresponding sequencing techniques |
1) 基因组DNA序列:全基因组测序[5](Whole genome sequencing,WGS),全外显子测序[6](Whole exome sequencing,WES)。
2) DNA甲基化修饰:全基因组重亚硫酸盐甲基化测序[7-9](Whole genome bisulfite sequencing,WGBS),氧化结合重亚硫酸氢盐测序[10](Oxidative bisulfite sequencing,oxBS-Seq)。
3) DNA-蛋白质相互作用:染色质免疫共沉淀测序[11-13](Chromatin immunoprecipitation sequencing,ChIP-Seq),染色质内源性切割与高通量测序[14](Chromatin endogenous cleavage with high-throughput sequencing,ChEC-Seq),CUT&Tag[15](Cleavage under targets and tagmentation)。
4) 染色质可及性:转座酶染色质可及性高通量测序分析[16](Assay for transposase-accessible chromatin with high throughput sequencing,ATAC-seq),微球菌核酸酶消化测序[17-18](Micrococcal nuclease digestion with deep sequencing,MNase-seq),脱氧核糖核酸酶I超敏位点测序[19](DNase I hypersensitive sites sequencing,DNase-seq),甲醛辅助分离调控元件[20](Formaldehyde-assisted isolation of regulatory elements,FAIRE-seq)。
5) DNA-DNA相互作用:基于染色体构象捕获(Chromosome conformation capture,3C)的测序技术,主要包括3C[21],4C[22-23],5C[24],HiC[25],与ChIP-Seq结合有HiChIP[26],ChIA-PET[27]等。
6) DNA-RNA相互作用:原位全RNA-DNA相互作用测序[28](in situ global RNA interactions with DNA by deep sequencing,GRID-seq),RNA-基因组原位定位[29](in situ mapping of RNA-genome interactome, iMARGI)。
1 染色质测序技术原理及应用 1.1 基因组DNA序列 1.1.1 WGSWGS是利用NGS基本原理针对全基因组进行测序,不同的测序平台使用的方法可能略有区别,但主要流程都包括DNA纯化(DNA purification)、DNA片段化(DNA shearing)、片段选择(Size selection)、末端修复(End repair)、加接头(Adaptor ligation)、PCR扩增(PCR amplification), 文库质控(Quality assessment of libraries)和上机测序(Sequencing)[30]。最终研究人员再通过数据分析[31-32]可以获得样本DNA的变异信息,包括点突变(Single nucleotide polymorphism,SNP)、短片段插入缺失(Small deletion and insertion,indel)、拷贝数变异(Copy number variations,CNVs)、结构变异(Structural variations,SVs)等,而获得这些突变信息则有助于研究者更好地理解与阐述肿瘤发生发展的基础机制。
1.1.2 WES外显子组约占基因组的1%,但85%疾病相关突变存在于外显子[33]。与WGS不同的是,WES只需设计外显子探针即可捕获外显子序列并进行高通量测序。相较于WGS,WES更经济,数据量更小,应用更加广泛,但其包含的信息不如WGS全面,非编码区域和内含子相关致病突变无法检出[34]。
1.1.3 应用目前,多种肿瘤类型和超过5万例的肿瘤基因组已完成了测序,研究人员并依此建立了多个肿瘤相关数据库并描绘出了各肿瘤的突变图谱,如TCGA[35], ICGC[36]和COSMIC[37]等。TCGA数据库覆盖超30个常见癌种,其中肺癌、乳腺癌、结直肠癌和血液病更是多达近万例,数据库提供包括基因组突变信息、拷贝数变化、表观遗传改变和基因表达谱等数据,在肿瘤分子分型、肿瘤特异标志物和药物靶向治疗等方面参考作用极大。ICGC是国际癌症基因组联合体,其目标是在全球范围全面阐述全球人类多达50种不同癌症类型中存在的基因组变化。截至目前,ICGC(Release 28)包含24 298个肿瘤基因组,检测到81 782 588个体细胞突变,涉及57 905个突变基因。COSMIC是世界上最大最全的关于肿瘤体细胞突变及其作用的数据库。最新版本(v97,2022-11-29)中涵盖超过150万个肿瘤样本的23 443 841个基因组变异, 16 015 511个非编码突变、19 428个融合突变, 321 804个基因组重排, 1 207 190个拷贝数变异,9 215 470个表达异常和7 930 489差异甲基化CpGs的详细信息。
研究表明,不同的肿瘤突变图谱表现不尽相同。例如,在乳腺癌中根据ER+/-与否,突变谱略有不同,但高频突变均主要为TP53, PIK3CA, MYC, ERBB2, CCND1, ZNF703/FGFR1,GATA3和RB1等[38-40]。而在胰腺导管腺癌中,KRAS突变比例则高达90%,其余靠前的有TP53, CDKN2A, SMAD4和BRCA1/2[41]等。笔者团队通过WES技术鉴定出ERBB2/3通路突变是胆囊癌的高频突变[42],此后在印度的一项研究中这一结果也进一步得到了证实[43],随后我们分别在细胞水平[44-45]和单细胞转录组水平[46]发现ERBB2/3突变与PD-L1, MDK表达相关,从而影响胆囊癌免疫微环境。进一步笔者团队还描绘了胆囊神经内分泌肿瘤的突变谱,其中高频突变为TP53, SNX27, PCDHB14, RB1,SRPK1和ZNF107等[47]。
通过对肿瘤驱动基因的突变情况进行描述,有助于肿瘤发生发展基础机制的研究和分子分型,最终实现靶向治疗、精准治疗[48-50]。
1.2 DNA甲基化实际上,除了DNA序列本身发生突变外,DNA还会发生许多化学修饰,目前已知的修饰至少有17种[51],其中以胞嘧啶(C)的5号碳原子甲基化(5-methylcytosine,5mC)修饰丰度最高,因此5mC也被称为第5种碱基。这些修饰能在不改变DNA序列的前提下引起DNA构象、DNA稳定性和DNA-蛋白质互作的改变,从而影响基因表达,在肿瘤进程中发挥重要作用。
1.2.1 WGBS5mC甲基化修饰大多发生在CpG双核苷酸,可在转录水平抑制基因表达。超过半数基因的启动子区域会有CpG的富集,这些区域称为CpG岛(CpG island,CpGi)[52]。WBGS的基本原理是利用重亚硫酸盐处理DNA,其中未被甲基化修饰的胞嘧啶(C)会脱氨基转换为尿嘧啶(U),再经后续的PCR扩增,U会转换为T,从而区分甲基化C和未甲基化C,最后结合高通量测序技术,即可获得单碱基分辨率的全基因组5mC修饰图谱[7-9, 53]。
1.2.2 oxBS-Seq5mC在TET酶作用下产生5-羟甲基胞嘧啶(5-hydroxymethylcytosine,5hmC),5hmC可能是活化DNA去甲基过程的中间产物,但同时也是一种稳定的DNA修饰,具体作用机制还不十分清楚,可能与DNA的主动/被动去甲基化有关[54]。WGBS无法区分5mC和5hmC,而oxBS-Seq首先将5hmC氧化为5-甲酰胞嘧啶(5-formylcytosine, 5fC),再经重亚硫酸盐处理则会转换为U,通过与BS-Seq的结果取差集即可实现5mC的精准检测和5hmC的检出[10, 53, 55],从而获得单碱基分辨率的5hmC修饰图谱。
1.2.3 应用DNA甲基化多起到抑制转录的作用,主要有影响转录因子与启动子结合、直接与转录抑制因子结合、影响组蛋白修饰和染色质结构等机制[56-57],而DNA甲基表观修饰的异常和失衡在肿瘤的发生和发展中发挥重要作用。例如:研究人员利用WGBS描绘了前列腺癌甲基化图谱,发现RXRG, FH, CCDC6,RARA和GPX3等抑癌基因呈高甲基化并低表达,而NCOA4, BIRC2,FGF6和HGF等癌基因呈低甲基化并高表达[57-58];苏建忠教授团队通过对210例早期乳腺癌患者的cfDNA甲基化情况进行分析,成功构建了一个新的早期乳腺癌预测模型,结合影像学可提高诊断的准确性[59];Berman B P等[60]研究人员发现与正常相比,结直肠癌包含更多低甲基化区域,而这些区域在局部表现为高甲基化但在长距离(>100 kb)下表现为低甲基化,并且这些区域与染色质高级结构相关。
DNA甲基化修饰癌症发生发展密切相关[61],利用WGBS等技术,研究人员可对DNA表观修饰图谱进行描绘[62],丰富肿瘤发生机制,为肿瘤早期诊断、治疗提供另一思路。
1.3 DNA-蛋白质相互作用染色质内的蛋白质包括组蛋白和非组蛋白,非组蛋白主要有转录因子(Transcription factors,TFs)、各类酶以及维持染色质拓扑结构的蛋白等。其中,组蛋白还可以发生多种修饰,如乙酰化、磷酸化、甲基化、苏木化、泛素化[63]等,这些修饰多见于H3的赖氨酸(K)和精氨酸(R)位点。蛋白质-DNA相互作用同样属于表观遗传学范畴,在转录调控、DNA复制、DNA损伤修复、DNA包装和修饰等方面发挥重要作用[64]。
1.3.1 ChIP-SeqChIP-Seq是ChIP和高通量测序结合的技术,首先利用甲醛交联蛋白质-DNA,随后分离DNA,并用超声波使其片段化,再通过特异性抗体与靶蛋白形成免疫复合物沉淀,最后解交联纯化得到目的DNA序列[11-13, 65]。再经测序和分析后,即可得到靶蛋白与目的DNA序列的互作图谱。
1.3.2 ChEC-Seq尽管ChIP-Seq已在研究蛋白质-DNA互作中发挥重要作用,但该技术仍存在一定的缺陷,主要原因在于使用了甲醛交联,甲醛处理会增加目的蛋白与其他可溶性蛋白的结合,从而增加非特异性信号[66]。ChEC-Seq是基于MNase的分析技术,首先需要构建靶蛋白-MNase融合蛋白,低Ca2+浓度下MNase不发挥作用,当添加合适的Ca2+后,MNase会切割靶蛋白两端的DNA序列,从而释放出目的序列[14, 67]。ChEC-Seq避免了甲醛交联和超声打断DNA的步骤,有效地规避了ChIP假阳性的问题,但该技术针对不同蛋白均需构建蛋白-MNase融合体,技术难度较高。
1.3.3 CUT&TagCUT&Tag是Henikoff博士研发的一种新的蛋白质-DNA互作分析方法[15],与ChIP-Seq相比,具有操作简便,细胞需求量低,背景噪音低等优点。其基本原理[15]是利用了预装DNA接头的protein A/G-Tn5转座酶融合蛋白,首先一抗与靶蛋白特异性结合,二抗放大信号,随后融合蛋白与二抗结合,通过加入Mg2+即可激活Tn5转座酶的活性,切割与靶蛋白结合的DNA序列并插入接头序列,最后分离标签化的DNA片段,即可用于建库测序。
1.3.4 应用基因组正常功能的发挥很大程度上依赖于动态的DNA-蛋白互作的调节,一旦这些平衡被打破,细胞内环境稳态将遭到破坏,从而引起病变甚至癌变。例如,Koeffler等[68]在食管癌细胞系中以H3K4me1,H3K4me3,H3K27ac以及转录因子等为靶标进行ChIP-Seq分析,鉴定了ALDH3A1超级增强子调控通路,TP63,SOX2和KLF5等转录因子起到关键调控作用;武汉大学吴旻教授[69]团队以H3K27ac, H3K4me3为靶标对73对结直肠癌标本进行ChIP-Seq分析,同时整合RNA-Seq和WGS数据,描绘了结直肠癌中增强子和超级增强子的改变情况,并在细胞系中验证了相关癌基因KLF3,MAFK和RUNX1等的生物学功能;Janssens等[70]利用CUT&Tag对急性髓系和淋巴系白血病的易位基因KMT2A进行了研究,发现KMT2A与HOX9,MEIS1和MEF2C等基因发生融合,融合蛋白可改变染色质组蛋白修饰模式,从而诱导致癌性染色质重排,促进白血病的发生。
DNA-蛋白质相互作用在染色质功能调节方面起到非常重要的作用,主要通过组蛋白修饰[71-72]和转录因子结合改变[73]等方式实现,描绘肿瘤的这些表观遗传变化将进一步阐明肿瘤发生发展机制,为肿瘤药物开发提供新的靶点。
1.4 染色质可及性染色质可及性是DNA可被启动子、增强子、绝缘子等顺式调控元件和转录因子、结构蛋白等蛋白质物理接近的性质。尽管可接近DNA仅占总DNA序列的2%~3%,但却包含90%以上的转录结合区域[74]。染色质的开放与关闭处于一个动态平衡的状态,其异常改变可能在肿瘤发生发展中发挥重要作用。
1.4.1 ATAC-SeqATAC-Seq主要原理[16, 74]是用预装接头的Tn5转座酶捕获开放染色质(Open chromatin)区域,Tn5可实现对基因组DNA的切割并将标签插入两端,闭合染色质(Closed chromatin)由于其结构致密而无法被切割,以此完成了对可及染色质序列的富集,随后对这些DNA序列进行高通量测序即可描绘出全基因组的染色质可及性图谱。ATAC-Seq操作简便,所需细胞量较少,但Tn5转座酶的价格较为昂贵,结果存在酶切割的序列偏好问题[75]。
1.4.2 MNase/DNase-SeqMNase具有核酸内切酶和外切酶的特性,优先靶向核小体之间的连接DNA(Linker DNA),使用MNase消化基因组DNA可实现对核小体序列的富集[17-18, 74],MNase-Seq主要用于核小体DNA的研究。DNase-Seq是使用DNase I切割开放染色质区域超敏位点,结合组蛋白或转录因子区域受到蛋白质的保护作用不会被切割,纯化切割后的DNA加接头序列,可进行高通量测序[19, 74, 76]。MNase/DNase-Seq原理简单,所需细胞量较多,且DNase I切割DNA时具有一定的偏好性,其分析的可靠性受到了一定的质疑,但MNase/DNase-Seq的结果,可间接推测出核小体以及转录因子的结合位点[75]。
1.4.3 FAIRE-SeqFAIRE-Seq的主要原理是缠绕组蛋白的核小体DNA与游离DNA在苯酚-氯仿中的溶解度不同。先用甲醛固定染色质后,再经超声打断,核小体DNA主要分布在有机相和水相之间,而游离DNA则分布于水相中,富集纯化游离DNA建库测序,以此完成对开放染色质的分析[20, 77]。FAIRE-Seq克服了MNase和DNase I的序列偏好性[75],但仍存在信噪比低的问题。各技术比较如图 2所示。
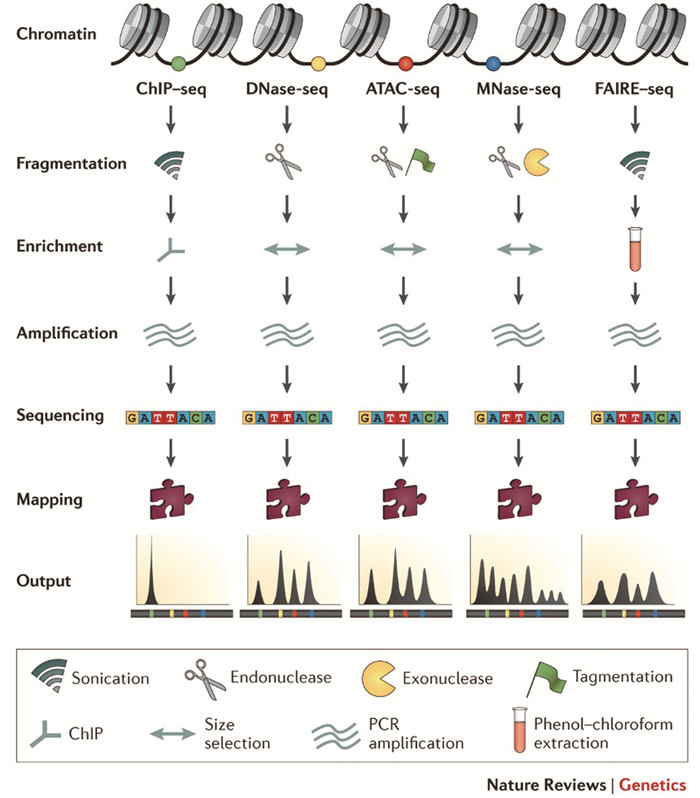
|
图 2 ChIP-Seq, DNase-Seq, ATAC-seq, MNase-Seq和FAIRE-Seq技术比较[75] Figure 2 An overview of ChIP-seq, DNase-seq, ATAC-seq, MNase-seq and FAIRE-seq experiments[75] |
目前关于肿瘤染色质可及性最大规模的研究来自于斯坦福大学Howard Y Chang教授和William J. Greenleaf教授研究团队[78]。作为ATAC-Seq的两位主要开发者,他们利用该技术绘制了来自TCGA样本库中的410例肿瘤样本涉及23种不同癌症类型的全基因组染色质可及性图谱,他们鉴定了562 709个可重复泛癌染色质可及性峰,其中约65%的泛癌峰与过往研究中发现的调控元件一致,表明ATAC-Seq不仅能很好地重复过去的研究,同时还能发现大量新的开放染色质位点。染色质开放/可及性是活性DNA的标志,通过结合基因组突变图谱,化学修饰图谱,转录谱等综合分析,可进一步描绘肿瘤基因调控网络,推动肿瘤基础机制的研究和肿瘤精准治疗。
1.5 DNA-DNA相互作用Job Dekker于2002年研发了染色体构象捕获(3C)技术[21],3D-基因组的研究自此拉开了序幕。随着后来基于3C的技术及测序方法不断开发与应用,染色质三维结构和染色质互作也逐渐被人们解析。基于3C的技术本质上检测的是DNA-DNA相互作用,如备受关注的增强子-启动子环(Enhancer-Promotor loop,E-P loop)。
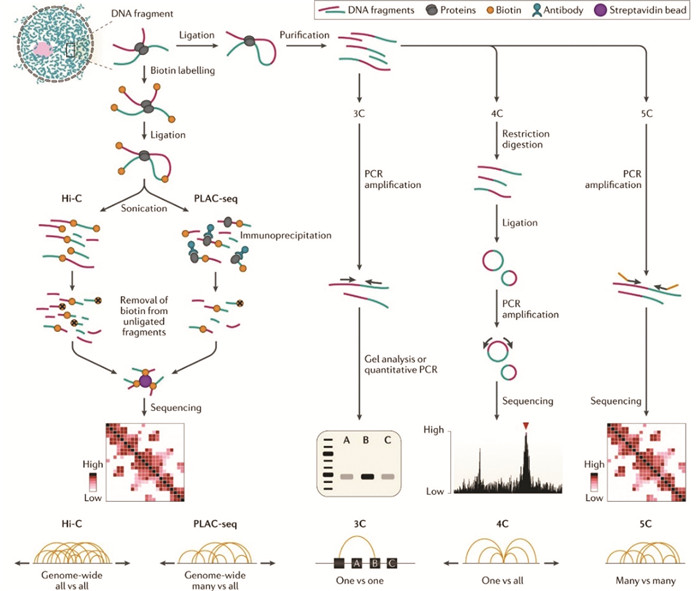
1.5.1 HiCHiC不同于3C,4C或5C的主要是因为3C是检测一对一(One vs one)的互作,4C是一对多(One vs all),5C是多对多(Many vs many),而HiC理论上可以检测所有(All vs all)的DNA互作[21-25, 79-81]。这些技术之所以被称为基于3C的技术, 是因为有着共同的处理步骤,主要包括甲醛固定、内切酶消化、连接、DNA纯化等步骤,如图 3所示。HiC不同的是首先用生物素标记的脱氧腺苷(Biotin labled dATP)填补了酶切末端,再连接成环,解交联后用超声将DNA打断,最后富集生物素标记的DNA片段建库测序,以此实现全基因组DNA-DNA相互作用的检测。
1.5.2 HiChIP/ChIA-PETHiChIP和ChIA-PET检出的DNA互作集合均是HiC的子集,主要用于分析特定蛋白相关的DNA互作。HiChIP是HiC和ChIP的结合,固定、酶切、连接后先用抗体富集靶蛋白-DNA复合物,随后再针对这些DNA建库测序[26]。ChIA-PET的原理略有不同,是ChIP与PET(Paired-end Tag sequencing)的结合,甲醛固定细胞后,超声打断DNA,随后特异抗体富集靶蛋白-DNA,接下来在DNA末端添加包含标签以及Mme I酶切位点生物素标记的linker DNA,连接后用Mme I消化并富集含生物素片段,即可测序。传统ChIA-PET只能检测2×20 bp的短序列,获得的信息量较少,近几年,有研究人员通过引入Tn5转座酶将检出序列扩大到2×250 bp,提高了DNA检出效率,同时也提高了覆盖SNP的可能[82]。
1.5.3 应用由于价格昂贵、技术复杂以及常常需要结合多种测序方法综合分析,基于HiC等技术的肿瘤染色质三维结构研究数量并不太多。Aristotelis等[83]通过对急性T淋巴细胞白血病原代细胞、细胞系以及正常T细胞进行比较,发现T-ALL与T细胞相比,其TAD内部的交互以及TAD边界的绝缘效应存在差异,而CTCF介导的TAD融合可能促进MYC启动子-超级增强子环的形成,从而促进肿瘤的形成。P. Andrew[84]通过对结肠癌、黑色素瘤、食管癌细胞系进行HiC建库,结合已发表的肿瘤TAD边界的数据并注释,再整合覆盖42种肿瘤类型超3 000例肿瘤-正常配对样本的WGS数据,发现肿瘤体细胞突变的分布受染色质三维结构的影响,TAD边界的突变分布率(20.6%)高于TAD内部(6.9%)。Bradley等[85]通过对结直肠癌临床样本以及细胞系样本进行了HiC建库,鉴定出超25 000个E-P环与肿瘤转录相关,同时提出在结直肠癌中除了A compartment和B compartment外还存在一种intermediate compartment的中间状态,极大地丰富了染色质三维结构理论模型。
同时,近年来研究人员发现染色质三维结构与结构变异和非编码序列改变密切相关,染色质的重排影响顺式调控元件(Cis-regulatory elements,CREs)从而激活癌基因的表达[86-88]。通过整合分析WGS、ChIP-seq和HiC等数据[89],可描绘出这种改变。笔者团队近期同样致力于胆道系统肿瘤和胰腺癌染色质三维结构的研究,有望描绘出上述肿瘤的染色质三维结构图谱。
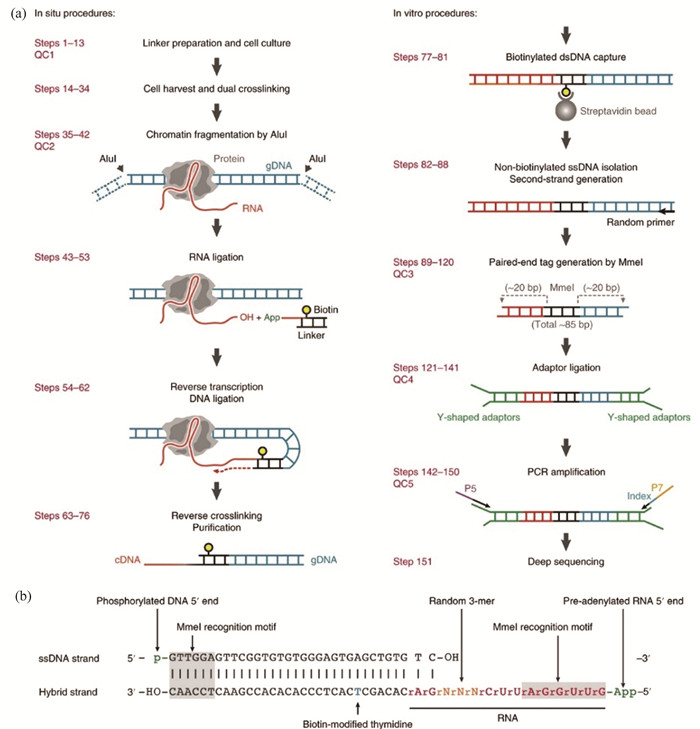
1.6 DNA-RNA相互作用 1.6.1 GRID-Seq自3-D基因组研究开展以来,DNA-DNA互作和RNA-RNA互作检测技术[90-91]相继得到了突破,但DNA-RNA互作检测技术迟迟未能突破。2017年,付向东教授团队研发出GRID-Seq可实现全基因组DNA-RNA互作的研究[28, 92]。其核心是设计了一段生物素标记的包含Mme I酶切位点的特殊的DNA-RNA linker,基本流程如图 4所示。首先用羟琥珀酰亚胺戊二酸(DSG)和甲醛双固定细胞内DNA-RNA互作,然后用Alu I消化基因组DNA,随后linker与RNA结合,再经连接和复制延长即可捕获DNA-RNA相互作用,最后经Mme I消化后,对约85 bp长的目的序列进行建库测序。
1.6.2 iMARGIiMARGI基本流程与GRID-Seq类似,同样需要一段特殊的linker捕获染色质上的RNA。与GRID-Seq不同的是,iMARGI中linker的酶切位点为BamH I,且需要环化捕获的DNA-RNA序列,经BamH I消化后,目的序列位于中间,两端为标签序列[29]。
1.6.3 应用目前,iMARGI还未在肿瘤研究中得到应用,而GRID-Seq除了付向东教授以三阴性乳腺癌细胞系MDA-MB-231为模型建立DNA-RNA互作图谱[28]外,其他类型肿瘤研究中也还未得到应用。他们在该细胞系中共检出868条高度富集的mRNA和72条ncRNA,可能在转录调控、稳定染色质构象等方面起到重要作用。DNA-RNA互作在肿瘤发生发展中的机制还需要以上技术在多种肿瘤类型中得到更多应用才能逐渐清晰。
2 国内研究成果得益于染色质测序技术的飞速发展以及成本的降低,国内众多肿瘤研究专家和团队对各类肿瘤展开了系统研究,由于成果众多,不胜枚举,笔者在此仅简单总结近十年国内团队利用临床样本研究的部分代表性成果(表 1)。由于肿瘤组织的异质性和复杂性,ChIP-Seq、和HiC等技术还未在临床样本中得到广泛应用,多数研究目前仍使用细胞系作为研究对象。若能够结合稳定的细胞分选(流式分选或磁珠分选)或类器官培养技术,使得样本的细胞类型尽可能均一,上述技术还有很大的应用空间。
| 表 1 国内利用染色质测序技术在肿瘤研究中的成果 Table 1 Achievements in cancer research using NGS-based chromatin sequencing in China |
值得注意的是,上述的大多测序技术在肿瘤单细胞研究[121-124]中已经得到了应用,推动了肿瘤异质性研究的快速发展。笔者也认为未来测序技术的发展趋势是精细化,单细胞化。随着单细胞分离技术和基因编辑技术的日渐成熟,CRISPR screening和单细胞测序的结合也孕育而生,目前已在转录组[125-127]和染色质可及性[128]方面得到了部分应用,CRISPR/Cas9基因编辑系统功能强大,与高通量测序有机结合潜力巨大。同时也应注意,尽管二代测序技术极大程度地推动了肿瘤研究的进展,但由于其读长较短(几百bp),在寻找串联重复(Tandem duplication)和大范围结构变异劣势较为明显[129]。近年来,基于Oxford nanopore和PacBio SMRT的单分子三代测序(Third generation sequencing,TGS)技术迅速发展,克服了二代测序读长短且需PCR扩增的缺点,可直接通过核酸分子读出长达数千bp的序列以及DNA/RNA的修饰信息,在寻找串联重复、结构变异和遗传信息从头组装等方面具有强大优势,但三代测序目前仍存在错误率较高、成本高昂和分析方法有限等问题[130-132]。由于本文主要探讨基于NGS的染色质测序技术,同时受于篇幅所限,单细胞染色质测序和三代测序技术的原理及应用在此不详细展开。
4 总结与展望染色质不仅储存着遗传信息,同时还有着精密的高级结构。针对肿瘤染色质分子生物学的不同方面,研究人员利用不同的基于NGS的染色质测序技术可对其进行研究,进而发现肿瘤染色质结构和功能的变化,并据此阐述肿瘤发生发展的基础机制,推动肿瘤的分子分型、靶向治疗和精准治疗。
| [1] |
FIERZ B, POIRIER M G. Biophysics of chromatin dynamics[J]. Annual Review of Biophysics, 2019, 48: 321-45. DOI:10.1146/annurev-biophys-070317-032847 (  0) 0) |
| [2] |
李国强, 刘法涛, 刘颖斌. 染色质三维结构变化在肿瘤进展中的作用[J]. 中华外科杂志, 2020, 58(12): 973-976. LI Guoqiang, LIU Fatao, LIU Yingbin. Role of three-dimensional chromatin structure changes in tumor progression[J]. Chinese Journal of Surgery, 2020, 58(12): 973-976. DOI:10.3760/cma.j.cn112139-20200319-00240 (  0) 0) |
| [3] |
TESSARZ P, KOUZARIDES T. Histone core modifications regulating nucleosome structure and dynamics[J]. Nature Reviews Molecular Cell Biology, 2014, 15(11): 703-708. DOI:10.1038/nrm3890 (  0) 0) |
| [4] |
MISTELI T. The self-organizing genome: Principles of genome architecture and function[J]. Cell, 2020, 183(1): 28-45. DOI:10.1016/j.cell.2020.09.014 (  0) 0) |
| [5] |
BENTLEY D R, BALASUBRAMANIAN S, SWERDLOW H P, et al. Accurate whole human genome sequencing using reversible terminator chemistry[J]. Nature, 2008, 456(7218): 53-59. DOI:10.1038/nature07517 (  0) 0) |
| [6] |
NG S B, TURNER E H, ROBERTSON P D, et al. Targeted capture and massively parallel sequencing of 12 human exomes[J]. Nature, 2009, 461(7261): 272-276. DOI:10.1038/nature08250 (  0) 0) |
| [7] |
MEISSNER A, MIKKELSEN T S, GU H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells[J]. Nature, 2008, 454(7205): 766-770. DOI:10.1038/nature07107 (  0) 0) |
| [8] |
LISTER R, O'MALLEY R C, TONTI-FILIPPINI J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis[J]. Cell, 2008, 133(3): 523-536. DOI:10.1016/j.cell.2008.03.029 (  0) 0) |
| [9] |
LISTER R, PELIZZOLA M, DOWEN R H, et al. Human DNA methylomes at base resolution show widespread epigenomic differences[J]. Nature, 2009, 462(7271): 315-322. DOI:10.1038/nature08514 (  0) 0) |
| [10] |
BOOTH M J, BRANCO M R, FICZ G, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution[J]. Science (New York, NY), 2012, 336(6083): 934-937. DOI:10.1126/science.1220671 (  0) 0) |
| [11] |
BARSKI A, CUDDAPAH S, CUI K, et al. High-resolution profiling of histone methylations in the human genome[J]. Cell, 2007, 129(4): 823-837. DOI:10.1016/j.cell.2007.05.009 (  0) 0) |
| [12] |
JOHNSON D S, MORTAZAVI A, MYERS R M, et al. Genome-wide mapping of in vivo protein-DNA interactions[J]. Science (New York, NY), 2007, 316(5830): 1497-1502. DOI:10.1126/science.1141319 (  0) 0) |
| [13] |
MIKKELSEN T S, KU M, JAFFE D B, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells[J]. Nature, 2007, 448(7153): 553-560. DOI:10.1038/nature06008 (  0) 0) |
| [14] |
ZENTNER G E, KASINATHAN S, XIN B, et al. ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo[J]. Nature Communications, 2015, 6: 8733. DOI:10.1038/ncomms9733 (  0) 0) |
| [15] |
KAYA-OKUR H S, WU S J, CODOMO C A, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells[J]. Nature Communications, 2019, 10(1): 1930. DOI:10.1038/s41467-019-09982-5 (  0) 0) |
| [16] |
BUENROSTRO J D, GIRESI P G, ZABA L C, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position[J]. Nature Methods, 2013, 10(12): 1213-1218. DOI:10.1038/nmeth.2688 (  0) 0) |
| [17] |
JOHNSON S M, TAN F J, MCCULLOUGH H L, et al. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin[J]. Genome Research, 2006, 16(12): 1505-1516. (  0) 0) |
| [18] |
SCHONES D E, CUI K, CUDDAPAH S, et al. Dynamic regulation of nucleosome positioning in the human genome[J]. Cell, 2008, 132(5): 887-898. DOI:10.1016/j.cell.2008.02.022 (  0) 0) |
| [19] |
BOYLE A P, DAVIS S, SHULHA H P, et al. High-resolution mapping and characterization of open chromatin across the genome[J]. Cell, 2008, 132(2): 311-322. DOI:10.1016/j.cell.2007.12.014 (  0) 0) |
| [20] |
GIRESI P G, KIM J, MCDANIELL R M, et al. FAIRE (Formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin[J]. Genome Research, 2007, 17(6): 877-885. DOI:10.1101/gr.5533506 (  0) 0) |
| [21] |
DEKKER J, RIPPE K, DEKKER M, et al. Capturing chromosome conformation[J]. Science (New York, NY), 2002, 295(5558): 1306-1311. DOI:10.1126/science.1067799 (  0) 0) |
| [22] |
ZHAO Z, TAVOOSIDANA G, SJÖLINDER M, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions[J]. Nature Genetics, 2006, 38(11): 1341-1347. DOI:10.1038/ng1891 (  0) 0) |
| [23] |
SIMONIS M, KLOUS P, SPLINTER E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C)[J]. Nature Genetics, 2006, 38(11): 1348-1354. DOI:10.1038/ng1896 (  0) 0) |
| [24] |
DOSTIE J, RICHMOND T A, ARNAOUT R A, et al. Chromosome conformation capture carbon copy (5C): A massively parallel solution for mapping interactions between genomic elements[J]. Genome Research, 2006, 16(10): 1299-1309. DOI:10.1101/gr.5571506 (  0) 0) |
| [25] |
LIEBERMAN-AIDEN E, VAN BERKUM N L, WILLIAMS L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome[J]. Science (New York, NY), 2009, 326(5950): 289-293. DOI:10.1126/science.1181369 (  0) 0) |
| [26] |
MUMBACH M R, RUBIN A J, FLYNN R A, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture[J]. Nature Methods, 2016, 13(11): 919-922. DOI:10.1038/nmeth.3999 (  0) 0) |
| [27] |
LI G, FULLWOOD M J, XU H, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing[J]. Genome Biology, 2010, 11(2): R22. DOI:10.1186/gb-2010-11-2-r22 (  0) 0) |
| [28] |
LI Xiao, ZHOU Bing, CHEN Liang, et al. GRID-seq reveals the global RNA-chromatin interactome[J]. Nature Biotechnology, 2017, 35(10): 940-950. DOI:10.1038/nbt.3968 (  0) 0) |
| [29] |
WU Weixin, YAN Zhangming, NGUYEN T C, et al. Mapping RNA-chromatin interactions by sequencing with iMARGI[J]. Nature Protocols, 2019, 14(11): 3243-3272. DOI:10.1038/s41596-019-0229-4 (  0) 0) |
| [30] |
YOSHINAGA Y, DAUM C, HE G, et al. Genome sequencing[J]. Methods In Molecular Biology (Clifton, NJ), 2018, 1775: 37-52. DOI:10.1007/978-1-4939-7804-5_4 (  0) 0) |
| [31] |
ULINTZ P J, WU W, GATES C M. Bioinformatics analysis of whole exome sequencing data[J]. Methods In Molecular Biology (Clifton, NJ), 2019, 1881: 277-318. DOI:10.1007/978-1-4939-8876-1_21 (  0) 0) |
| [32] |
KOSUGI S, MOMOZAWA Y, LIU X, et al. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing[J]. Genome Biology, 2019, 20(1): 117. DOI:10.1186/s13059-019-1720-5 (  0) 0) |
| [33] |
CHOI M, SCHOLL U I, JI W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(45): 19096-19101. DOI:10.1073/pnas.0910672106 (  0) 0) |
| [34] |
NAKAGAWA H, FUJITA M. Whole genome sequencing analysis for cancer genomics and precision medicine[J]. Cancer Science, 2018, 109(3): 513-522. DOI:10.1111/cas.13505 (  0) 0) |
| [35] |
Cancer Genome Atlas Research Network. The cancer genome atlas[DB/OL]. (2008-09-04)[2020-12-01]. https://www.cancer.gov/ccg/research/genome-sequencing/tcga.
(  0) 0) |
| [36] |
HUDSON T J, ANDERSON W, ARTEZ A, et al. International network of cancer genome projects[J]. Nature, 2010, 464(7291): 993-998. DOI:10.1038/nature08987 (  0) 0) |
| [37] |
TATE J G, BAMFORD S, JUBB H C, et al. COSMIC: the catalogue of somatic mutations in cancer[J]. Nucleic Acids Research, 2019, 47(D1): D941-D947. DOI:10.1093/nar/gky1015 (  0) 0) |
| [38] |
STEPHENS P J, TARPEY P S, DAVIES H, et al. The landscape of cancer genes and mutational processes in breast cancer[J]. Nature, 2012, 486(7403): 400-404. DOI:10.1038/nature11017 (  0) 0) |
| [39] |
PEREIRA B, CHIN S-F, RUEDA O M, et al. The somatic mutation profiles of 2, 433 breast cancers refines their genomic and transcriptomic landscapes[J]. Nature Communications, 2016, 7: 11479. DOI:10.1038/ncomms11479 (  0) 0) |
| [40] |
NIK-ZAINAL S, DAVIES H, STAAF J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences[J]. Nature, 2016, 534(7605): 47-54. DOI:10.1038/nature17676 (  0) 0) |
| [41] |
WADDELL N, PAJIC M, PATCH A M, et al. Whole genomes redefine the mutational landscape of pancreatic cancer[J]. Nature, 2015, 518(7540): 495-501. DOI:10.1038/nature14169 (  0) 0) |
| [42] |
LI Maolan, ZHANG Zhou, LI Xiaoguang, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway[J]. Nature Genetics, 2014, 46(8): 872-876. DOI:10.1038/ng.3030 (  0) 0) |
| [43] |
PANDEY A, STAWISKI E W, DURINCK S, et al. Integrated genomic analysis reveals mutated ELF3 as a potential gallbladder cancer vaccine candidate[J]. Nature Communications, 2020, 11(1): 4225. DOI:10.1038/s41467-020-17880-4 (  0) 0) |
| [44] |
GENG Yajun, CHEN Shili, YANG Yang, et al. Long-term exposure to genistein inhibits the proliferation of gallbladder cancer by downregulating the MCM complex[J]. Science Bulletin, 2022, 67(8): 813-824. DOI:10.1016/j.scib.2022.01.011 (  0) 0) |
| [45] |
LI Maolan, LIU Fatao, ZHANG Fei, et al. Genomic ERBB2/ERBB3 mutations promote PD-L1-mediated immune escape in gallbladder cancer: A whole-exome sequencing analysis[J]. Gut, 2019, 68(6): 1024-1033. DOI:10.1136/gutjnl-2018-316039 (  0) 0) |
| [46] |
ZHANG Yijian, ZUO Chunman, LIU Liguo, et al. Single-cell RNA-sequencing atlas reveals an MDK-dependent immunosuppressive environment in ErbB pathway-mutated gallbladder cancer[J]. Journal of Hepatology, 2021, 75(5): 1128-1141. DOI:10.1016/j.jhep.2021.06.023 (  0) 0) |
| [47] |
LIU Fatao, LI Yongsheng, YING Dongjian, et al. Whole-exome mutational landscape of neuroendocrine carcinomas of the gallbladder[J]. Signal Transduction and Targeted Therapy, 2021, 6(1): 55. DOI:10.1038/s41392-020-00412-3 (  0) 0) |
| [48] |
NEGRINI S, GORGOULIS V G, HALAZONETIS T D. Genomic instability-an evolving hallmark of cancer[J]. Nature Reviews Molecular Cell Biology, 2010, 11(3): 220-228. DOI:10.1038/nrm2858 (  0) 0) |
| [49] |
HUANG A, GARRAWAY L A, ASHWORTH A, et al. Synthetic lethality as an engine for cancer drug target discovery[J]. Nature Reviews Drug Discovery, 2020, 19(1): 23-38. DOI:10.1038/s41573-019-0046-z (  0) 0) |
| [50] |
ZHURAVLEVA E, O'ROURKE C J, ANDERSEN J B. Mutational signatures and processes in hepatobiliary cancers[J]. Nature Reviews Gastroenterology & Hepatology, 2022, 19(6): 367-382. DOI:10.1038/s41575-022-00587-w (  0) 0) |
| [51] |
RAIBER E A, HARDISTY R, VAN DELFT P, et al. Mapping and elucidating the function of modified bases in DNA[J]. Nature Reviews Chemistry, 2017, 1(9): 0069. DOI:10.1038/s41570-017-0069 (  0) 0) |
| [52] |
JONES P A. Functions of DNA methylation: islands, start sites, gene bodies and beyond[J]. Nature Reviews Genetics, 2012, 13(7): 484-492. DOI:10.1038/nrg3230 (  0) 0) |
| [53] |
ZHAO Linyong, SONG Jinghui, LIU Yibin, et al. Mapping the epigenetic modifications of DNA and RNA[J]. Protein & Cell, 2020, 11(11): 792-808. DOI:10.1007/s13238-020-00733-7 (  0) 0) |
| [54] |
WU Xiaoji, ZHANG Yi. TET-mediated active DNA demethylation: Mechanism, function and beyond[J]. Nature Reviews Genetics, 2017, 18(9): 517-534. DOI:10.1038/nrg.2017.33 (  0) 0) |
| [55] |
BOOTH M J, OST T W B, BERALDI D, et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine[J]. Nature Protocols, 2013, 8(10): 1841-1851. DOI:10.1038/nprot.2013.115 (  0) 0) |
| [56] |
DAS P M, SINGAL R. DNA methylation and cancer[J]. Journal of Clinical Oncology : Official Journal of the American Society of Clinical Oncology, 2004, 22(22): 4632-4642. DOI:10.1200/JCO.2004.07.151 (  0) 0) |
| [57] |
YU Y P, DING Ying, CHEN Rui, et al. Whole-genome methylation sequencing reveals distinct impact of differential methylations on gene transcription in prostate cancer[J]. The American Journal of Pathology, 2013, 183(6): 1960-1970. DOI:10.1016/j.ajpath.2013.08.018 (  0) 0) |
| [58] |
ZHAO S G, CHEN W S, LI Haolong, et al. The DNA methylation landscape of advanced prostate cancer[J]. Nature Genetics, 2020, 52(8): 778-789. DOI:10.1038/s41588-020-0648-8 (  0) 0) |
| [59] |
LIU Jiaqi, ZHAO Hengqiang, HUANG Yukuan, et al. Genome-wide cell-free DNA methylation analyses improve accuracy of non-invasive diagnostic imaging for early-stage breast cancer[J]. Molecular Cancer, 2021, 20(1): 36. DOI:10.1186/s12943-021-01330-w (  0) 0) |
| [60] |
BERMAN B P, WEISENBERGER D J, AMAN J F, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains[J]. Nature Genetics, 2011, 44(1): 40-46. DOI:10.1038/ng.969 (  0) 0) |
| [61] |
KOCH A, JOOSTEN S C, FENG Z, et al. Analysis of DNA methylation in cancer: Location revisited[J]. Nature Reviews Clinical Oncology, 2018, 15(7): 459-466. DOI:10.1038/s41571-018-0004-4 (  0) 0) |
| [62] |
PAPANICOLAU-SENGOS A, ALDAPE K. DNA methylation profiling: an emerging paradigm for cancer diagnosis[J]. Annual Review of Pathology, 2022, 17: 295-321. DOI:10.1146/annurev-pathol-042220-022304 (  0) 0) |
| [63] |
ZHANG Yanjun, SUN Zhongxing, JIA Junqi, et al. Overview of Histone Modification[J]. Advances In Experimental Medicine and Biology, 2021, 1283: 1-16. DOI:10.1007/978-981-15-8104-5_1 (  0) 0) |
| [64] |
FERRAZ R A C, LOPES A L G, DA SILVA J A F, et al. DNA-protein interaction studies: a historical and comparative analysis[J]. Plant Methods, 2021, 17(1): 82. DOI:10.1186/s13007-021-00780-z (  0) 0) |
| [65] |
YAMAKAWA A, HOJO H, OHBA S. ChIP-Seq assays from mammalian cartilage and chondrocytes[J]. Methods In Molecular Biology (Clifton, NJ), 2021, 2245: 167-178. DOI:10.1007/978-1-0716-1119-7_12 (  0) 0) |
| [66] |
BARANELLO L, KOUZINE F, SANFORD S, et al. ChIP bias as a function of cross-linking time[J]. Chromosome Research : An International Journal on the Molecular, Supramolecular and Evolutionary Aspects of Chromosome Biology, 2016, 24(2): 175-181. DOI:10.1007/s10577-015-9509-1 (  0) 0) |
| [67] |
SCHMID M, DURUSSEL T, LAEMMLI U K. ChIC and ChEC; genomic mapping of chromatin proteins[J]. Molecular Cell, 2004, 16(1): 147-157. DOI:10.1016/j.molcel.2004.09.007 (  0) 0) |
| [68] |
JIANG Yanyi, JIANG Yuan, LI Chunquan, et al. TP63, SOX2, and KLF5 establish a core regulatory circuitry that controls epigenetic and transcription patterns in esophageal squamous cell carcinoma cell lines[J]. Gastroenterology, 2020, 159(4): 1311-1327.e19. DOI:10.1053/j.gastro.2020.06.050 (  0) 0) |
| [69] |
LI Qinglan, LIN Xiang, YU Yali, et al. Genome-wide profiling in colorectal cancer identifies PHF19 and TBC1D16 as oncogenic super enhancers[J]. Nature Communications, 2021, 12(1): 6407. DOI:10.1038/s41467-021-26600-5 (  0) 0) |
| [70] |
JANSSENS D H, MEERS M P, WU S J, et al. Automated CUT&Tag profiling of chromatin heterogeneity in mixed-lineage leukemia[J]. Nature Genetics, 2021, 53(11): 1586-1596. DOI:10.1038/s41588-021-00941-9 (  0) 0) |
| [71] |
MILLáN-ZAMBRANO G, BURTON A, BANNISTER A J, et al. Histone post-translational modifications - cause and consequence of genome function[J]. Nature Reviews Genetics, 2022, 23(9): 563-580. DOI:10.1038/s41576-022-00468-7 (  0) 0) |
| [72] |
ZHU Dongwei, ZHANG Yue, WANG Shengjun. Histone citrullination: A new target for tumors[J]. Molecular Cancer, 2021, 20(1): 90. DOI:10.1186/s12943-021-01373-z (  0) 0) |
| [73] |
BUSHWELLER J H. Targeting transcription factors in cancer-from undruggable to reality[J]. Nature Reviews Cancer, 2019, 19(11): 611-624. DOI:10.1038/s41568-019-0196-7 (  0) 0) |
| [74] |
KLEMM S L, SHIPONY Z, GREENLEAF W J. Chromatin accessibility and the regulatory epigenome[J]. Nature Reviews Genetics, 2019, 20(4): 207-220. DOI:10.1038/s41576-018-0089-8 (  0) 0) |
| [75] |
MEYER C A, LIU X S. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology[J]. Nature Reviews Genetics, 2014, 15(11): 709-721. DOI:10.1038/nrg3788 (  0) 0) |
| [76] |
SONG Lingyun, CRAWFORD G E. DNase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells[J]. Cold Spring Harbor Protocols, 2010, 2010(2): 5384. DOI:10.1101/pdb.prot5384 (  0) 0) |
| [77] |
GIRESI P G, LIEB J D. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements)[J]. Methods (San Diego, Calif), 2009, 48(3): 233-239. DOI:10.1016/j.ymeth.2009.03.003 (  0) 0) |
| [78] |
CORCES M R, GRANJA J M, SHAMS S, et al. The chromatin accessibility landscape of primary human cancers[J]. Science (New York, NY), 2018, 362: 6413. DOI:10.1126/science.aav1898 (  0) 0) |
| [79] |
DE WIT E, DE LAAT W. A decade of 3C technologies: Insights into nuclear organization[J]. Genes & Development, 2012, 26(1): 11-24. DOI:10.1101/gad.179804.111 (  0) 0) |
| [80] |
KEMPFER R, POMBO A. Methods for mapping 3D chromosome architecture[J]. Nature Reviews Genetics, 2020, 21(4): 207-226. DOI:10.1038/s41576-019-0195-2 (  0) 0) |
| [81] |
NOORDERMEER D, DUBOULE D. Chromatin looping and organization at developmentally regulated gene loci[J]. Wiley Interdisciplinary Reviews Developmental Biology, 2013, 2(5): 615-630. DOI:10.1002/wdev.103 (  0) 0) |
| [82] |
LI Xingwang, LUO O J, WANG Ping, et al. Long-read ChIA-PET for base-pair-resolution mapping of haplotype-specific chromatin interactions[J]. Nature Protocols, 2017, 12(5): 899-915. DOI:10.1038/nprot.2017.012 (  0) 0) |
| [83] |
KLOETGEN A, THANDAPANI P, NTZIACHRISTOS P, et al. Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia[J]. Nature Genetics, 2020, 52(4): 388-400. DOI:10.1038/s41588-020-0602-9 (  0) 0) |
| [84] |
AKDEMIR K C, LE V T, KIM J M, et al. Somatic mutation distributions in cancer genomes vary with three-dimensional chromatin structure[J]. Nature Genetics, 2020, 52(11): 1178-1188. DOI:10.1038/s41588-020-0708-0 (  0) 0) |
| [85] |
JOHNSTONE S E, REYES A, QI Y, et al. Large-scale topological changes restrain malignant progression in colorectal cancer[J]. Cell, 2020, 182(6): 1474-1489.e23. DOI:10.1016/j.cell.2020.07.030 (  0) 0) |
| [86] |
SPIELMANN M, LUPIáñEZ D G, MUNDLOS S. Structural variation in the 3D genome[J]. Nature Reviews Genetics, 2018, 19(7): 453-467. DOI:10.1038/s41576-018-0007-0 (  0) 0) |
| [87] |
DUBOIS F, SIDIROPOULOS N, WEISCHENFELDT J, et al. Structural variations in cancer and the 3D genome[J]. Nature Reviews Genetics, 2022, 22(9): 533-546. DOI:10.1038/s41568-022-00488-9 (  0) 0) |
| [88] |
KHURANA E, FU Y, CHAKRAVARTY D, et al. Role of non-coding sequence variants in cancer[J]. Nature Reviews Genetics, 2016, 17(2): 93-108. DOI:10.1038/nrg.2015.17 (  0) 0) |
| [89] |
WANG Xiaotao, XU Jie, ZHANG Baozhen, et al. Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes[J]. Nature Methods, 2021, 18(6): 661-668. DOI:10.1038/s41592-021-01164-w (  0) 0) |
| [90] |
KUDLA G, GRANNEMAN S, HAHN D, et al. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(24): 10010-10015. DOI:10.1073/pnas.1017386108 (  0) 0) |
| [91] |
CAI Zhaokui, CAO Changchang, JI Lei, et al. RIC-seq for global in situ profiling of RNA-RNA spatial interactions[J]. Nature, 2020, 582(7812): 432-437. DOI:10.1038/s41586-020-2249-1 (  0) 0) |
| [92] |
ZHOU Bing, LI Xiao, LUO Daji, et al. GRID-seq for comprehensive analysis of global RNA-chromatin interactions[J]. Nature Protocols, 2019, 14(7): 2036-2068. DOI:10.1038/s41596-019-0172-4 (  0) 0) |
| [93] |
FANG Wenfeng, JIN Haoxuan, ZHOU Haoqiang, et al. Intratumoral heterogeneity as a predictive biomarker in anti-PD-(L)1 therapies for non-small cell lung cancer[J]. Molecular Cancer, 2021, 20: 37. DOI:10.1186/s12943-021-01331-9 (  0) 0) |
| [94] |
WANG Cheng, DAI Juncheng, QIN Na, et al. Analyses of rare predisposing variants of lung cancer in 6, 004 whole genomes in Chinese[J]. Cancer Cell, 2022, 40(10): 1223-1239.e6. DOI:10.1016/j.ccell.2022.08.013 (  0) 0) |
| [95] |
XUE Ruidong, LI Ruoyan, GUO Hua, et al. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma[J]. Gastroenterology, 2016, 150(4): 998-1008. DOI:10.1053/j.gastro.2015.12.033 (  0) 0) |
| [96] |
XU L X, HE M H, DAI Z H, et al. Genomic and transcriptional heterogeneity of multifocal hepatocellular carcinoma[J]. Annals of Oncology : Official Journal of the European Society For Medical Oncology, 2019, 30(6): 990-997. DOI:10.1093/annonc/mdz103 (  0) 0) |
| [97] |
LIU Weiwei, ZHENG Lu, ZHANG Rongguiyi, et al. Circ-ZEB1 promotes PIK3CA expression by silencing miR-199a-3p and affects the proliferation and apoptosis of hepatocellular carcinoma[J]. Molecular Cancer, 2022, 21: 72. DOI:10.1186/s12943-022-01529-5 (  0) 0) |
| [98] |
ZHOU Shaolai, ZHOU Zhengjun, SONG Chengli, et al. Whole-genome sequencing reveals the evolutionary trajectory of HBV-related hepatocellular carcinoma early recurrence[J]. Signal Transduction and Targeted Therapy, 2022, 7: 24. DOI:10.1038/s41392-021-00838-3 (  0) 0) |
| [99] |
DONG Liangqing, SHI Yang, MA Lijie, et al. Spatial and temporal clonal evolution of intrahepatic cholangiocarcinoma[J]. Journal of Hepatology, 2018, 69(1): 89-98. DOI:10.1016/j.jhep.2018.02.029 (  0) 0) |
| [100] |
DONG Liangqing, LU Dayun, CHEN Ran, et al. Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma[J]. Cancer Cell, 2022, 40(1): 70-87.e15. DOI:10.1016/j.ccell.2021.12.006 (  0) 0) |
| [101] |
SONG Yongmei, LI Lin, OU Yunwei, et al. Identification of genomic alterations in oesophageal squamous cell cancer[J]. Nature, 2014, 509(7498): 91-95. DOI:10.1038/nature13176 (  0) 0) |
| [102] |
GAO Yibo, CHEN Zhaoli, LI Jiagen, et al. Genetic landscape of esophageal squamous cell carcinoma[J]. Nature Genetics, 2014, 46(10): 1097-1092. DOI:10.1038/ng.3076 (  0) 0) |
| [103] |
ZHANG Ling, ZHOU Yong, CHENG Caixia, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma[J]. American Journal of Human Genetics, 2015, 96(4): 597-611. DOI:10.1016/j.ajhg.2015.02.017 (  0) 0) |
| [104] |
CUI Yongping, CHEN Hongyan, XI Ruibin, et al. Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma[J]. Cell Research, 2020, 30(10): 902-913. DOI:10.1038/s41422-020-0333-6 (  0) 0) |
| [105] |
HONG Xiafei, QIAO Sitan, LI Fuqiang, et al. Whole-genome sequencing reveals distinct genetic bases for insulinomas and non-functional pancreatic neuroendocrine tumours: Leading to a new classification system[J]. Gut, 2020, 69(5): 877-887. DOI:10.1136/gutjnl-2018-317233 (  0) 0) |
| [106] |
JIANG Lu, GU Zhaohui, YAN Zixun, et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma[J]. Nature Genetics, 2015, 47(9): 1061-1066. DOI:10.1038/ng.3358 (  0) 0) |
| [107] |
HUANG Liang, LIU Dan, WANG Na, et al. Integrated genomic analysis identifies deregulated JAK/STAT-MYC-biosynthesis axis in aggressive NK-cell leukemia[J]. Cell Research, 2018, 28(2): 172-186. DOI:10.1038/cr.2017.146 (  0) 0) |
| [108] |
XIONG Jie, CUI Bowen, WANG Nan, et al. Genomic and transcriptomic characterization of natural killer T cell lymphoma[J]. Cancer Cell, 2020, 37(3): 403-419.e6. DOI:10.1016/j.ccell.2020.02.005 (  0) 0) |
| [109] |
HUANG Yaohui, CAI Kun, XU Pengpeng, et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis[J]. Signal Transduction and Targeted Therapy, 2021, 6: 10. DOI:10.1038/s41392-020-00437-8 (  0) 0) |
| [110] |
GUO Guangwu, SUN Xiaojuan, CHEN Chao, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation[J]. Nature Genetics, 2013, 45(12): 1459-1463. DOI:10.1038/ng.2798 (  0) 0) |
| [111] |
REN Shancheng, WEI Gonghong, LIU Dongbing, et al. Whole-genome and transcriptome sequencing of prostate cancer identify new genetic alterations driving disease progression[J]. European Urology, 2018, 73(3): 322-339. DOI:10.1016/j.eururo.2017.08.027 (  0) 0) |
| [112] |
HU Zheng, ZHU Da, WANG Wei, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism[J]. Nature Genetics, 2015, 47(2): 158-163. DOI:10.1038/ng.3178 (  0) 0) |
| [113] |
HE Juan, ZENG Zhen, WANG Yuelong, et al. Characterization of novel CTNNB1 mutation in Craniopharyngioma by whole-genome sequencing[J]. Molecular Cancer, 2021, 20(1): 168. DOI:10.1186/s12943-021-01468-7 (  0) 0) |
| [114] |
MA Gaoxiang, LIU Hanting, HUA Qiuhan, et al. KCNMA1 cooperating with PTK2 is a novel tumor suppressor in gastric cancer and is associated with disease outcome[J]. Molecular Cancer, 2017, 16(1): 46. DOI:10.1186/s12943-017-0613-z (  0) 0) |
| [115] |
ZUO Ying, ZHONG Jia, BAI Hua, et al. Genomic and epigenomic profiles distinguish pulmonary enteric adenocarcinoma from lung metastatic colorectal cancer[J]. EBioMedicine, 2022, 82: 104165. DOI:10.1016/j.ebiom.2022.104165 (  0) 0) |
| [116] |
LI Juan, LIANG Yuan, FAN Jian, et al. DNA methylation subtypes guiding prognostic assessment and linking to responses the DNA methyltransferase inhibitor SGI-110 in urothelial carcinoma[J]. BMC Medicine, 2022, 20(1): 222. DOI:10.1186/s12916-022-02426-w (  0) 0) |
| [117] |
ZHANG Yuanyuan, CHEN Hongyan, MO Hongnan, et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer[J]. Cancer Cell, 2021, 39(12): 1578-1593.e8. DOI:10.1016/j.ccell.2021.09.010 (  0) 0) |
| [118] |
WANG Huanjun, MEI Yan, LUO Cheng, et al. Single-cell analyses reveal mechanisms of cancer stem cell maintenance and epithelial-mesenchymal transition in recurrent bladder cancer[J]. Clinical Cancer Research : An Official Journal of the American Association for Cancer Research, 2021, 27(22): 6265-6278. DOI:10.1158/1078-0432.CCR-20-4796 (  0) 0) |
| [119] |
LIANG Zhengyu, LI Guipeng, WANG Zejun, et al. BL-Hi-C is an efficient and sensitive approach for capturing structural and regulatory chromatin interactions[J]. Nature Communications, 2017, 8(1): 1622. DOI:10.1038/s41467-017-01754-3 (  0) 0) |
| [120] |
YANG Lu, CHEN Fengling, ZHU Haichuan, et al. 3D genome alterations associated with dysregulated HOXA13 expression in high-risk T-lineage acute lymphoblastic leukemia[J]. Nature Communications, 2021, 12(1): 3708. DOI:10.1038/s41467-021-24044-5 (  0) 0) |
| [121] |
WU S J, FURLAN S N, MIHALAS A B, et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression[J]. Nature Biotechnology, 2021, 39(7): 819-824. DOI:10.1038/s41587-021-00865-z (  0) 0) |
| [122] |
TAAVITSAINEN S, ENGEDAL N, CAO S, et al. Single-cell ATAC and RNA sequencing reveal pre-existing and persistent cells associated with prostate cancer relapse[J]. Nature Communications, 2021, 12(1): 5307. DOI:10.1038/s41467-021-25624-1 (  0) 0) |
| [123] |
HOU Yu, GUO Huahu, CAO Chen, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas[J]. Cell Research, 2016, 26(3): 304-319. DOI:10.1038/cr.2016.23 (  0) 0) |
| [124] |
GROSSELIN K, DURAND A, MARSOLIER J, et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer[J]. Nature Genetics, 2019, 51(6): 1060-1066. DOI:10.1038/s41588-019-0424-9 (  0) 0) |
| [125] |
DATLINGER P, RENDEIRO A F, SCHMIDL C, et al. Pooled CRISPR screening with single-cell transcriptome readout[J]. Nature Methods, 2017, 14(3): 297-301. DOI:10.1038/nmeth.4177 (  0) 0) |
| [126] |
ADAMSON B, NORMAN T M, JOST M, et al. A multiplexed single-cell crispr screening platform enables systematic dissection of the unfolded protein response[J]. Cell, 2016, 167(7): 1867-1882.e21. DOI:10.1016/j.cell.2016.11.048 (  0) 0) |
| [127] |
REPLOGLE J M, NORMAN T M, XU A, et al. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing[J]. Nature Biotechnology, 2020, 38(8): 954-961. DOI:10.1038/s41587-020-0470-y (  0) 0) |
| [128] |
RUBIN A J, PARKER K R, SATPATHY A T, et al. Coupled single-cell crispr screening and epigenomic profiling reveals causal gene regulatory networks[J]. Cell, 2019, 176(1-2): 361-376.e17. DOI:10.1016/j.cell.2018.11.022 (  0) 0) |
| [129] |
KUMAR K R, COWLEY M J, DAVIS R L. Next-generation sequencing and emerging technologies[J]. Seminars In Thrombosis and Hemostasis, 2019, 45(7): 661-673. DOI:10.1055/s-0039-1688446 (  0) 0) |
| [130] |
SEARLE B, MüLLER M, CARELL T, et al. Third-generation sequencing of epigenetic DNA[J]. Angewandte Chemie (International Ed In English), 2022, 62(14): e202215704. DOI:10.1002/anie.202215704 (  0) 0) |
| [131] |
JUNG H, WINEFIELD C, BOMBARELY A, et al. Tools and strategies for long-read sequencing and de novo assembly of plant genomes[J]. Trends In Plant Science, 2019, 24(8): 700-724. DOI:10.1016/j.tplants.2019.05.003 (  0) 0) |
| [132] |
PETERSEN L M, MARTIN I W, MOSCHETTI W E, et al. Third-generation sequencing in the clinical laboratory: Exploring the advantages and challenges of nanopore sequencing[J]. Journal of Clinical Microbiology, 2019, 58(1): e01315-19. DOI:10.1128/JCM.01315-19 (  0) 0) |
 2024, Vol. 22
2024, Vol. 22