表观遗传学是研究可逆、可遗传的表型变化,不涉及核DNA序列的变化[1]。虽然表观遗传学的全部范围尚未确定,但其一般含义是指化学修饰主要包括DNA、RNA甲基化,组蛋白修饰以及非编码RNA修饰和染色质重排。在表观遗传修饰中,DNA甲基化和组蛋白修饰得到了很好的研究,例如DNA中的5-甲基胞嘧啶甲基化已经在多种肿瘤中影响着基因表达。近些年的对于甲基化药物的研究已经取得明显的进展,如去甲基化药物地西他滨和阿扎胞苷,以及组蛋白去乙酰化酶抑制剂塞达明,为治疗临床疾病提供了更多策略[2-3]。而对于RNA,已经在生物体中鉴定了100多种RNA的修饰[4],RNA甲基化是RNA修饰的主要形式,广泛存在于各种类型的RNA中,如rRNA、tRNA、snRNA、snoRNA以及mRNA中。特别是mRNA的N6-甲基腺苷(N6-methyladenosine,m6A)修饰是哺乳动物基因表达的必需调节因子[5-6]。其他修饰如5-甲基胞嘧啶(5-methylcytosine,m5C)和N1-甲基腺苷(N1-methyladenosine,m1A)[7]。其中,m6A修饰约占mRNA修饰的80%,近年来广泛被关注和研究。RNA m6A修饰是可逆的,其生物学效应主要通过“编码器”,“擦码器”和“读码器”蛋白介导[8]。编码器是将甲基化修饰“写入”RNA的过程,即介导RNA的甲基化修饰的过程,包括METTL3(Methyltransferase-like 3)、METTL14(Methyltransferase-like 14)、WTAP(Wilms tumor suppressor-1-associated protein)和ZC3H13(Zinc finger CCCH-type containing 13)[9]。擦码器可以“擦除”RNA甲基化修饰信号,即介导RNA的去甲基化过程,包括FTO(Fat mass and obesity-associated protein)和ALKBH5(Alpha-ketoglutarate-dependent dioxygenase homolog 5)[10-11]。读码器负责“读取” RNA甲基化信息并参与下游RNA的翻译和降解,包括YTHDC1(YTH domain-containing 1)、YTHDC2(YTH domain-containing 2)、YTHDF1(YTH N6-methyladenosine RNA binding protein 1)、YTHDF2(YTH N6-methyladenosine RNA binding protein 2)和HNRNPC(Heterogeneous nuclear ribonucleoprotein)[12]。越来越多的体外和体内研究揭示了m6A甲基化修饰在调节多种生物过程中的起到关键的作用,包括精子发生[13]、神经发生[14]、性别决定[15-16]等。异常m6A修饰在不同类型癌症的进展中起着至关重要的作用[17],尤其是作为乳腺癌[18]、胶质母细胞瘤[19]和白血病的癌干细胞的调节剂[20]。异常的m6A修饰对癌症的生长和进展至关重要,这表明m6A修饰途径可能是癌症治疗的一个有前途的治疗靶点。
肺癌是肿瘤相关死亡的主要原因,5年生存率仅为16.6%[21]。非小细胞肺癌(Non-small cell lung cancer,NSCLC)占肺癌的80%。在NSCLC中,两种主要亚型是肺腺癌(Lung adenocarcinoma,LUAD)和肺鳞状细胞癌(Lung squamous cell carcinoma,LUSC),分别占病例的50%-60%和30%。越来越多的研究表明,异常的m6A甲基化在包括肺癌在内的许多疾病的发病机制中起着至关重要的作用。去年Li[22]等人通过TCGA(The Cancer Genome Atlas)数据库来探讨m6A调节剂分子与肺腺癌的关系,本研究与Li等人的研究部分相类似,同样也探讨了m6A甲基化分子的表达情况。但与之不同的是,接下来的研究是按照各个分子是否与生存相关进行。为证实研究的可靠性,对GEO(Gene Expression Omnibus)中的数据进行分析来进一步验证,从而试图揭示肺腺癌发生和发展的分子机制,有助于助开发新的有效靶向治疗方案。
1 材料和方法 1.1 RNA-seq数据和生物信息学分析从TCGA肺腺癌癌项目官方网站(https://portal.gdc.cancer.gov/repository)下载基因表达数据(535例肿瘤组织和59例正常肺组织,数据类型:HTSeq-FPKM)和相应的临床信息。排除正常肺织样本,使用箱形图显示离散变量的表达差异[23]。最后,522例肺腺癌患者和临床资料)的RNA-Seq基因表达水平3 HTSeq FPKM数据并进一步分析。
1.2 统计分析用limma软件包分析了535例肿瘤组织和59例正常肺组织中12个编码已知m6A RNA甲基化调节因子的基因的表达情况(FC>1、p < 0.05;logFC < 0的差异表达基因为下调基因,log FC >0的差异表达基因为上调基因)。提取这12个基因的表达矩阵和与样品相关的临床信息。使用R中的pheatmap、vioplot和corrplot软件包生成热图、小提琴图和表达相关图。使用Kaplan-Meier方法分别计算12个m6A调节剂表达与肺腺癌患者的生存曲线。使用Cox回归与TCGA患者的总体存活相关的临床病理学特征。使用危险比(HRs)超过1表示为危险基因,而HRs < 1表示保护基因。Cox回归分析用于比较HNRNPC/YTHDF2的表达对生存率的影响以及其他临床特征(年龄、分期、组织浸润程度、淋巴结状态、远处转移状态)。最后,采用kruskal(KS)检验和logistic回归评估临床因素与HNRNPC表达之间的关联。缺失的临床数据通过列表删除进行处理;如果缺少任何参数的值,则整个样本将从分析中排除。OS的定义是从诊断日期到死亡日期的时间间隔。基因高、低表达是基于中位数确定的。所有统计分析均使用R软件(V.3.5.1)进行。
1.3 使用基因表达综合数据库(GEO)进行验证为了确保TCGA队列结果的准确性,使用GEO数据库(https://www.ncbi.nlm.nih.gov/geo/)中的GSE68465验证HNRNPC表达水平。数据集包括443个肺腺癌和19个相邻的非肿瘤组织样本。以及来验证HNRNPC是否是肺腺癌预后的独立预测因子。
2 结果分析 2.1 TCGA队列中肺腺癌患者的临床病理学特征2020年6月从TCGA数据库下载了522个具有临床和基因表达数据的原发性肿瘤(见表 1)。诊断时的中位年龄为67岁。研究发现临床Ⅰ期有279例(54.3%),Ⅱ期124例(24.1%),Ⅲ期85例(16.5%)和Ⅳ期26例(5.1%)。组织浸润为T1的有172例(33.1%),T2的有281例(54.1%),T3的有47例(9.1%),T4的有19例(3.7%)。510例中有175例(34.3%)有淋巴结转移。378例中有25例(6.7%)有远处转移。中位随访时间为18.3个月(范围0~227个月)。
| 表 1 肺腺癌患者的临床病理学特征 Table 1 Clinicopathological features of lung adenocarcinoma |
考虑每种RNA m6A甲基化调节剂在肿瘤发生和发展中的重要生物学功能。首先,比较了TCGA数据库中535个肺腺癌组织和59个正常肺组织中12种RNA m6A甲基化调节因子的表达水平。与正常肺组织相比,其中在肺腺癌组织中HNRNPC、YTHDF3、METTL3、YTHDF1、YTHDF2是相对高表达的,WTAP、FTO、ZC3H13、METTL14是低表达(见图 1a~1b,表 2,p < 0.05)。不同RNA m6A甲基化调节剂的相关比率从弱到强相关(见图 1c)。大多数RNA m6A甲基化调节因子之间的关系呈是正相关的,但是FTO和HNRNPC是呈现出负相关的。在这些基因中METTL14和YTHDC1基因最为相关,即当METTL14基因表达上调时、YTHDC1基因表达很有可能上调(见图 1c)。

|
图 1 RNA m6A甲基化相关基因在肺腺癌和正常肺组织中的表达 Figure 1 Expression of RNA m6A methylation regulators in lung adenocarcinoma and normal tissues 注:(a)表达越高或越低,颜色越暗(红色上调,绿色下调) *p < 0.05, **p < 0.01, ***p < 0.001;(b)小提琴图可视化肺腺癌中差异表的RNA m6A甲基化调节因子(蓝色是正常组织,红色是肺腺癌);(c)肺腺癌12个RNA m6A修饰调节因子的Spearman相关分析(红色代表正相关,蓝色代表负相关). |
| 表 2 RNA m6A调节剂的表达 Table 2 Expression of RNA m6A regulators |
除去正常肺组织对照组后,绘制肺腺癌中所有差异表达的m6A甲基化调节因子的生存曲线。发现仅仅YTHDF2和HNRNPC的表达与肺腺癌患者生存相关,即YTHDF2的高表达有更好的生存、HNRNPC低表达有更好的生存(见图 2)。
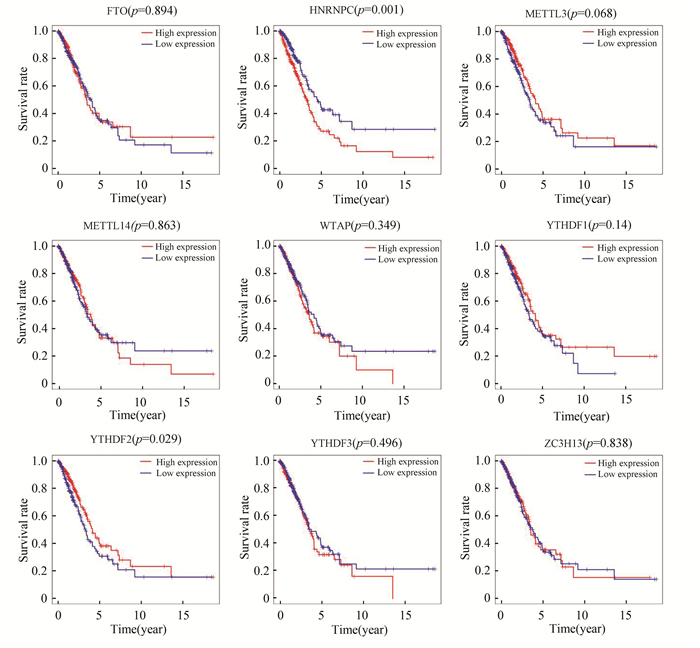
|
图 2 TCGA队列中所有差异表达的m6A甲基化调节因子的生存曲线 Figure 2 Survival curve of all differentially-expressed m6A methylation regulators in TCGA cohort |
采用单因素Cox回归分析显示HNRNPC的表达与总体生存相关([HR]: 1.012 046 411;95%CI: 1.000 432 8-1.023 794 84;p=0.042)(见表 3)。与生存相关的其他临床病理变量包括临床分期、组织浸润深度和淋巴结转移(见表 3)。虽然在前文所述Kaplan-Meier生存分析表明,YTHDF2高表达组预后好于YTHDF2低表达组(p=0.029),但是通过单因素Cox回归结果表明YTHDF2的表达与肺腺癌患者生存关系不大(见表 4)。
| 表 3 使用Cox回归的TCGA患者与总体存活和临床病理学特征的关联及变量选择后的多因素生存模型(HNRNPC) Table 3 Association of TCGA patients with overall survival and clinicopathological features using Cox regression (HNRNPC) and multivariate survival model after variable selection (HNRNPC) |
| 表 4 使用Cox回归的TCGA患者与总体存活和临床病理学特征的关联及变量选择后的多因素生存模型(YTHDF2) Table 4 Association of TCGA patients with overall survival and clinicopathological features using Cox regression and multivariate survival model after variable selection (YTHDF2) |
随后对这两个基因行多因素Cox回归分析,结果表明仅仅HNRNPC的表达以及临床分期可以作为生存独立的预后因子(HR=1.012 369 230, 95%CI: 1.000 351 911-1.024 530 914,p=0.044)(见表 3)、(见图 3),即HNRNPC在肺腺癌中可能作为独立危险预后因子。

|
图 3 Multivariate分析模型(森林图) Figure 3 Multivariate analysis model (Forest map) |
HNRNPC的表达与性别(p=0.02)、临床分期(p=0.026)、远处转移(p=0.014)和组织浸润深度(p=0.001)显著相关(见图 4)。使用logistic回归分析显示HNRNPC在肺腺癌中的表达与临床分期(Ⅳ vs Ⅰ, OR=3.692 308)和组织浸润深度(T2 vs T1, OR=1.776 471; T4 vs T1, OR=6.303 030)显著相关(见表 5, p < 0.05)。这些结果表明在肺腺癌,HNRNPC的表达水平倾向于与临床分期和组织浸润深度相关。

|
图 4 HNRNPC表达和临床病理特征的关系(Kruskal(KS)检验) Figure 4 Relationship between HNRNPC expression and clinicopathological features(Kruskal (KS) test) |
使用GSE68465进行生存分析提示HNRNPC低表达组有更好的生存(p=0.009,见图 5a)。单因素Cox分析显示HNRNPC的高表达与肺腺癌患者较低的总生存率相关,HR为1.367(95%CI:1.038-1.801,P=0.026)(见图 5b)。多因素Cox分析表明,HNRNPC表达水平是独立的预后因素,HR为1.393(95%CI: 1.060-1.831,p=0.017)(见图 5c)。通过验证后结果与之前TCGA数据库一致, 两次结果表明HNRNPC在肺腺癌中可能作为独立危险预后因子。
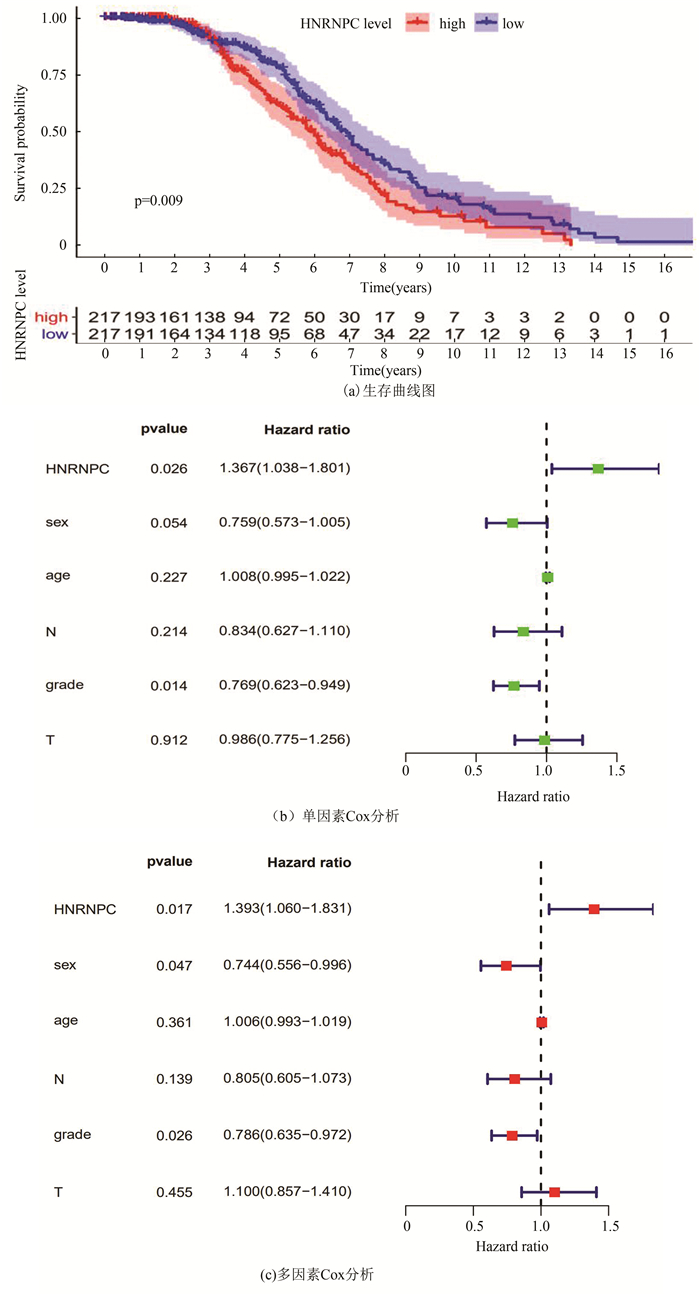
|
图 5 采用GEO进行数据验证(GSE68465) Figure 5 Validation by GEO (GSE68465) |
表观遗传学近年来已成为一个热门话题,m6A甲基化修饰是100多种不同RNA修饰中最丰富的化学修饰。1974年m6A修饰最初是在poly(A)RNA部分中被鉴定,并且预测它可能在mRNA加工中起作用[24]。有证据表明RNA m6A修饰与肿瘤增殖、分化、肿瘤发生[25]、增殖[26]、侵袭[25]和转移[27]有关。m6A修饰调节基因可能作为原癌基因或抑癌基因在肿瘤中发挥重要作用。
癌症是一种复杂的遗传和表观遗传疾病,涉及癌基因和抑癌基因的突变和失调[28]。其中,表观遗传修饰可以通过抑制抑癌基因的表达和增强癌基因的表达来参与肿瘤的发生和发展[29]。近年来,肺癌领域m6A修饰的研究逐渐进入研究者们的视野。Chao等[30]认为m6A去甲基化酶ALKBH5通过下调FOXM1 mRNA的m6A修饰水平和促进FOXM1的表达,影响间歇性缺氧条件下肺腺癌细胞的增殖和侵袭。同样作为擦码器,FTO通过降低肺鳞状细胞癌中MZF1 mRNA转录物中的m6A甲基化水平和mRNA稳定性来增强MZF1表达,从而起到肿瘤启动子的作用[31]。FTO可以通过上调USP7的表达来促进NSCLC细胞的生长[32]。对于相关阅读蛋白的研究,已经有研究结果表明,YTHDF2通过促进6-磷酸葡萄糖酸脱氢酶mRNA的翻译来促进肺癌细胞的生长[33]。因此在肺癌中,m6A相关基因和蛋白表达水平有可能成为肿瘤分子诊断的潜在标志物,也将为临床上分子靶向治疗的发展提供新的靶点。本研究是从生物信息学的角度发掘m6A甲基化因子在肺腺癌中的作用。
近期,Gao[34]等人从TCGA数据库中女性肺腺癌队列进行研究,该研究结果有助于评估女性肺腺癌患者的预后提供重要的理论支持。本研究首先评估了m6A调控相关基因表达谱,发现大多数m6A调节因子在肺腺癌中差异表达。其中HNRNPC、YTHDF3、METTL3、YTHDF1、YTHDF2表达上调;WTAP、FTO、ZC3H13、METTL14下调。然后探索了这些分子之间的内在联系,发现METTL14和YTHDC1是相关性最大的。也就是说,当METTL14高表达时,YTHDC1很可能表达上调。随后对这些差异表达的基因进行生存分析,Kaplan-Meier生存分析显示仅仅HNRNPC和YTHDF2的表达与总体存活相关。然后,通过multivariate分析显示HNRNPC可以成为肺腺癌总生存期的独立预后因素。随后,使用logistic regression回归的单因素分析显示HNRNPC在肺腺癌中的表达与临床分期(Ⅳ vs Ⅰ, OR=3.692 308)、组织浸润深度(T2 vs T1, OR=1.776 471;T4 vs T1, OR=6.303 03)(p < 0.05)显著相关。最后通过GEO数据库进行验证, 最后得出HNRNPC可能成为肺腺癌的独立预后因素。
在多种肿瘤或肿瘤细胞系中观察到HNRNPC的异常上调,包括胶质母细胞瘤[35],肝细胞癌[36],黑素瘤[37]和肺癌[38]。作为RNA结合蛋白(RNA binding protein,RBP),HNRNPC参与RNA剪接[39-40]、非特异性RNA输出[41]、RNA表达[35, 42]、稳定性[43-44]、3'末端加工[45]和翻译[46]。敲除HNRNPC可以减少细胞增殖并增强依托泊苷诱导的细胞凋亡,这表明HNRNPC在GBM中起着致癌基因的作用[35]。同样,HNRNPC过表达与胃癌细胞的化疗耐药密切相关,高表达水平的HNRNPC与临床预后不良有关[47]。KHSRP和HNRNPC可通过激活IFN-α-JAK-STAT1信号通路诱导人肺癌细胞侵袭和转移[48]。最近,Huang[49]等人发现HNRNPC的过表达通过上皮-间质转化(epithelial-mesenchymal transition,EMT)促进口腔鳞状细胞的癌变。在我们的研究中,本研究发现HNRNPC在肺腺癌组织中表达上调。然后进一步发现HNRNPC的表达水平与肺腺癌患者的总生存期相关,且与临床分期和组织浸润深度相关。最后发现HNRNPC可能作为独立的预后因素,从而可能为肺腺癌提供新的治疗靶点。
4 结论肺腺癌和正常对照之间发生的变化的RNA m6A甲基化调节因子,可能在肺腺癌的进展中起着至关重要的作用。HNRNPC表达可能是肺腺癌的潜在预后分子标志物,但是需要更多的研究进一步验证。
| [1] |
NG R K, GURDON J B. Epigenetic inheritance of cell differentiation status[J]. Cell Cycle, 2008(9): 1173-1177. DOI:10.4161/cc.7.9.5791 (  0) 0) |
| [2] |
WOUTERS B J, DELWEL R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia[J]. Blood, 2016, 127(1): 42-52. DOI:10.1182/blood-2015-07-604512 (  0) 0) |
| [3] |
LI Y, CHEN K, ZHOU Y, et al. A new strategy to target acute myeloid leukemia stem and progenitor cells using chidamide, a histone deacetylase inhibitor[J]. Current Cancer Drug Targets, 2015, 15(6): 493-503. DOI:10.2174/156800961506150805153230 (  0) 0) |
| [4] |
BOCCALETTO P, MACHNICKA M A, PURTA E, et al. MODOMICS: A database of RNA modification pathways 2017 update[J]. Nucleic Acids Research, 2018, 46(D1): D303-D307. DOI:10.1093/nar/gkx1030 (  0) 0) |
| [5] |
ROUNDTREE I A, EVANS M E, PAN T, et al. Dynamic RNA modifications in gene expression regulation[J]. Cell, 2017, 169(7): 1187-1200. DOI:10.1016/j.cell.2017.05.045 (  0) 0) |
| [6] |
GEULA S, MOSHITCH-MOSHKOVITZ S, DOMINISSINI D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation[J]. Science, 2015, 347(6225): 1002-1006. DOI:10.1126/science.1261417 (  0) 0) |
| [7] |
FRYE M, BLANCO S. Post-transcriptional modifications in development and stem cells[J]. Development, 2016, 143(21): 3871-3881. DOI:10.1242/dev.136556 (  0) 0) |
| [8] |
LIU N, PAN T. N6-methyladenosine-encoded epitranscriptomics[J]. Nature Structural & Molecular Biology, 2016, 23(2): 98-102. DOI:10.1038/nsmb.3162 (  0) 0) |
| [9] |
SCHOLLER E, WEICHMANN F, TREIBER T, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex[J]. RNA, 2018(4): 499-512. DOI:10.1261/rna.064063.117 (  0) 0) |
| [10] |
TANG C, KLUKOVICH R, PENG H, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells[J]. Proceedings of the National Academy of Sciences of the United States of Americ, 2018, 115(2): E325-e33. DOI:10.1073/pnas.1717794115 (  0) 0) |
| [11] |
DING C, ZOU Q, DING J, et al. Increased N6-methyladenosine causes infertility is associated with FTO expression. Journal of cellular physiology[J]. Journal of Cellular Physiology, 2018, 233(9): 7055-7066. DOI:10.1002/jcp.26507 (  0) 0) |
| [12] |
WOJTAS M N, PANDEY R R, MENDEL M, et al. Regulation of m(6)A transcripts by the 3'-->5' RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline[J]. Molecular Cell, 2017, 68(2): 374-387. DOI:10.1016/j.molcel.2017.09.021 (  0) 0) |
| [13] |
ZHENG G, DAHL J A, NIU Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility[J]. Molecular Cell, 2013, 49(1): 18-29. DOI:10.1016/j.molcel.2012.10.015 (  0) 0) |
| [14] |
YOON K J, RINGELING F R, VISSERS C, et al. Temporal control of mammalian cortical neurogenesis by m6A methylation[J]. Cell, 2017, 171(4): 877-889. DOI:10.1016/j.cell.2017.09.003 (  0) 0) |
| [15] |
HAUSSMANN IU, BODI Z, SANCHEZ-MORAN E, et al. M6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination[J]. Nature, 2016, 540(7632): 301-304. DOI:10.1038/nature20577 (  0) 0) |
| [16] |
LENCE T, AKHTAR J, BAYER M, et al. m6A modulates neuronal functions and sex determination in Drosophila[J]. Nature, 2016, 540(7632): 242-247. DOI:10.1038/nature20568 (  0) 0) |
| [17] |
HUANG H, WENG H, CHEN J. m6A Modification in coding and non-coding RNAs: Roles and therapeutic implications in cancer[J]. Cancer Cell, 2020, 37(3): 270-288. DOI:10.1016/j.ccell.2020.02.004 (  0) 0) |
| [18] |
ZHANG C, SAMANTA D, LU H, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(14): E2047-56. DOI:10.1073/pnas.1602883113 (  0) 0) |
| [19] |
CUI Q, SHI H, YE P, et al. m6A RNA Methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells[J]. Cell Reports, 2017, 18(11): 2622-2634. DOI:10.1016/j.celrep.2017.02.059 (  0) 0) |
| [20] |
WENG H, HUANG H, WU H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification[J]. Cell Stem Cell, 2018, 22(2): 191-205. DOI:10.1016/j.stem.2017.11.016 (  0) 0) |
| [21] |
CHEN W, ZHENG R, BAADE P D, et al. Cancer statistics in China, 2015[J]. A Cancer Journal for Clinicians, 2016, 66: 115e132. DOI:10.3322/caac.21338 (  0) 0) |
| [22] |
LI F, WANG H, HUANG H, et al. m6A RNA methylation regulators participate in the malignant progression and have clinical prognostic value in lung adenocarcinoma[J]. Frontiers in Genetics, 2020, 11: 994. DOI:10.3389/fgene.2020.00994 (  0) 0) |
| [23] |
KRUPPA J, JUNG K. Automated multigroup outlier identification in molecular highthroughput data using bagplots and gemplots[J]. BMC Bioinformatics, 2017, 18(1): 232. DOI:10.1186/s12859-017-1645-5 (  0) 0) |
| [24] |
DESROSIERS R, FRIDERICI K, ROTTMAN F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells[J]. Proceedings of the National Academy of Sciences of the United States of America, 1974, 71(10): 3971-3975. DOI:10.1073/pnas.71.10.3971 (  0) 0) |
| [25] |
LIN S, CHOE J, DU P, et al. The m6A methyltransferase METTL3 promotes translation in human cancer cells[J]. Molecular Cell, 2016, 62(3): 335-345. DOI:10.1016/j.molcel.2016.03.021 (  0) 0) |
| [26] |
LIU J, ECKERT M A, HARADA B T, et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer[J]. Nature Cell Biology, 2018, 20(9): 1074-1083. DOI:10.1038/s41556-018-0174-4 (  0) 0) |
| [27] |
MA J Z, YANG F, ZHOU C C, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary MicroRNA processing[J]. Hepatology, 2017, 65(2): 529-543. DOI:10.1002/hep.28885 (  0) 0) |
| [28] |
MAFFICINI A, SCARPA A. Genetics and epigenetics of gastroenteropancreatic neuroendocrine neoplasms[J]. Endocrine Reviews, 2019, 40(2): 506-536. DOI:10.1210/er.2018-00160 (  0) 0) |
| [29] |
KLUTSTEIN M, NEJMAN D, GREENFIELD R, et al. DNA methylation in cancer and aging[J]. Cancer Research, 2016, 76(12): 3446-3450. DOI:10.1158/0008-5472 (  0) 0) |
| [30] |
CHAO Y, SHANG J, JI W. ALKBH5-m6A-FOXM1 signaling axis promotes proliferation and invasion of lung adenocarcinoma cells under intermittent hypoxia[J]. Biochemical and Biophysical Research Communications, 2020, 521(2): 499-506. DOI:10.1016/j.bbrc.2019.10.145 (  0) 0) |
| [31] |
LIU J, REN D, DU Z, et al. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression[J]. Biochemical and Biophysical Research Communications, 2018, 502(4): 456-464. DOI:10.1016/j.bbrc.2018.05.175 (  0) 0) |
| [32] |
LI J, HAN Y, ZHANG H, et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA[J]. Biochemical and Biophysical Research Communications, 2019, 512(3): 479-485. DOI:10.1016/j.bbrc.2019.03.093 (  0) 0) |
| [33] |
SHENG H, LI Z, SU S, et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation[J]. Carcinogenesis, 2020, 41(5): 541-550. DOI:10.1093/carcin/bgz152 (  0) 0) |
| [34] |
GAO C, ZHUANG J, LI H, et al. Gene signatures of 6-methyladenine regulators in women with lung adenocarcinoma and development of a risk scoring system: A retrospective study using the cancer genome atlas database[J]. Aging (Albany NY), 2021, 13(3): 3957-3968. DOI:10.18632/aging.202364 (  0) 0) |
| [35] |
PARK Y M, HWANG S J, MASUDA K, et al. Heterogeneous nuclear ribonucleoprotein C1/C2 controls the metastatic potential of glioblastoma by regulating PDCD4[J]. Molecular and Cellular Biology, 2012, 32(20): 4237-4244. DOI:10.1128/MCB.00443-12 (  0) 0) |
| [36] |
SUN W, XING B, SUN Y, et al. Proteome analysis of hepatocellular carcinoma by two-dimensional difference gel electrophoresis: Novel protein markers in hepatocellular carcinoma tissues[J]. Molecular & Cellular Proteomics, 2007, 6(10): 1798-1808. DOI:10.1074/mcp.M600449-MCP200 (  0) 0) |
| [37] |
MULNIX R E, PITMAN R T, RETZER A, et al. hnRNP C1/C2 and Pur-beta proteins mediate induction of senescence by oligonucleotides homologous to the telomere overhang[J]. OncoTargets and Therapy, 2013, 7: 23-32. DOI:10.2147/OTT.S54575 (  0) 0) |
| [38] |
PINO I, PIO R, TOLEDO G, et al. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer[J]. Lung Cancer, 2003, 41(2): 131-143. DOI:10.1016/s0169-5002(03)00193-4 (  0) 0) |
| [39] |
KONIG J, ZARNACK K, ROT G, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution[J]. Nature Structural & Molecular Biology, 2010, 17(7): 909-915. DOI:10.1038/nsmb.1838 (  0) 0) |
| [40] |
ZARNACK K, KONIG J, TAJNIK M, et al. Direct competition between hnRNPC and U2AF65 protects the transcriptome from the exonization of Alu elements[J]. Cell, 2013, 152(3): 453-466. DOI:10.1016/j.cell.2012.12.023 (  0) 0) |
| [41] |
MCCLOSKEY A, TANIGUCHI I, SHINMYOZU K, et al. hnRNP C tetramer measures RNA length to classify RNA polymerase Ⅱ transcripts for export[J]. Science, 2012, 335: 1643-1646. DOI:10.1126/science.1218469 (  0) 0) |
| [42] |
CHRISTIAN K J, LANG M A, RAFFALLI-MATHIEU F. Interaction of heterogeneous nuclear ribonucleoprotein C1/C2 with a novel cis-regulatory element within p53 mRNA as a response to cytostatic drug treatment[J]. Molecular Pharmacology, 2008, 73(5): 1558-1567. DOI:10.1124/mol.107.042507 (  0) 0) |
| [43] |
SHETTY S. Regulation of urokinase receptor mRNA stability by hnRNP C in lung epithelial cells[J]. Molecular And Cellular Biochemistry, 2005, 272(1/2): 107-118. DOI:10.1007/s11010-005-7644-2 (  0) 0) |
| [44] |
VELUSAMY T, SHETTY P, BHANDARY Y P, et al. Posttranscriptional regulation of urokinase receptor expression by heterogeneous nuclear ribonuclear protein C[J]. Biochemistry, 2008, 47(24): 6508-6517. DOI:10.1021/bi702338y (  0) 0) |
| [45] |
GRUBER A J, SCHMIDT R, GRUBER A R, et al. A comprehensive analysis of 3' end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation[J]. Genome Research, 2016, 26(8): 1145-1159. DOI:10.1101/gr.202432.115 (  0) 0) |
| [46] |
LEE E K, KIM H H, KUWANO Y, et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies[J]. Nature Structural & Molecular Biology, 2010, 17(6): 732-739. DOI:10.1038/nsmb.1815 (  0) 0) |
| [47] |
HUANG H, HAN Y, ZHANG C, et al. HNRNPC as a candidate biomarker for chemoresistance in gastric cancer[J]. Tumor Biology, 2016, 37: 3527-3534. DOI:10.1007/s13277-015-4144-1 (  0) 0) |
| [48] |
YAN M, SUN L, LI J, et al. RNA-binding protein KHSRP promotes tumor growth and metastasis in non-small cell lung cancer[J]. Journal of Experimental & Clinical Cancer Research, 2019, 38(1): 478. DOI:10.1186/s13046-019-1479-2 (  0) 0) |
| [49] |
HUANG G Z, WU Q Q, ZHENG Z N, et al. m6A-related bioinformatics analysis reveals that HNRNPC facilitates progression of OSCC via EMT[J]. Aging (Albany NY), 2020, 12(12): 11667-11684. DOI:10.18632/aging.103333 (  0) 0) |
 2022, Vol. 20
2022, Vol. 20


