2. 甘肃农业大学 动物医学院,兰州 730070
2. College of Veterinary Medicine, Gansu Agricultural University, Lanzhou 730070, China
脑红蛋白(Neuroglobin,NGB)是德国科学家Burmester于2000年首次发现的一种特异性携氧球蛋白,主要分布于中枢神经系统[1]。与血红蛋白、肌红蛋白相比,NGB具有经典螺旋三折叠结构的特点,其基本结构是蛋白质肽链和1个血红蛋白[2]。蛋白质结构是生物长期演化出的结果,从DNA编码转录翻译到蛋白质序列折叠形成稳定的空间结构,并非一个随机产生的过程。对任意蛋白质的序列进行修改,即使只增加、删除或修改蛋白质序列中的一个氨基酸,蛋白质结构都会发生明显的改变,相应的蛋白质功能也会随之变化,而如果修改两个及以上氨基酸,多数蛋白将无法进行有效的折叠[3]。要想深入探究蛋白质的内部功能机制,则首先需要证明蛋白质结构与其功能之间的关系。近年来,国内外学者主要对NGB转录调控机制及其在神经元中的保护作用开展了相关研究工作[4-7],至于NGB蛋白结构学方面的研究,则多集中于对其基本结构[8]及功能相关核心位点的研究[9-10],国内目前对于该项工作的研究尚处于生物信息学的初步分析阶段[11],而针对NGB蛋白三级结构的详细预测分析还未见相关报道。本项目组李盛杰等[12]研究发现,天祝白牦牛NGB基因编码的氨基酸序列与普通黄牛相比,在第83、97位分别发生了丝氨酸(Ser)→脯氨酸(Pro)、精氨酸(Arg)→谷氨酰胺(Gln)的突变,但其生物学意义目前尚不明确。针对天祝白牦牛NGB蛋白两处氨基酸位点突变,本文拟从蛋白质三级结构模型分析突变造成的NGB蛋白结构变化,并与黄牛NGB蛋白的三级结构进行比较,推测天祝白牦牛NGB蛋白位点突变对其三级结构的影响,为进一步探究高原低氧环境下牦牛NGB蛋白的遗传特性和生理机制提供理论参考。
1 材料与方法 1.1 NGB蛋白同源性比较分析登陆Genbank(http://www.ncbi.nlm.nih.gov/genbank)下载天祝白牦牛、黄牛、甘南牦牛、绵羊、猕猴、人类、红原鸡等25个物种的NGB氨基酸序列(登录信息见表 1所示)。利用Clustal-Raindy、MEGA等软件对25个物种NGB蛋白氨基酸序列进行同源性比较,并建立系统进化树。
| 表 1 不同物种NGB蛋白序列信息 Table 1 Sequence information of NGB protein in different species |
利用Swiss Model(https://swissmodel.expasy.org/interactive)在线软件对天祝白牦牛、黄牛NGB蛋白进行三维建模,并用Spdbv软件进行蛋白质模型输出。
2 结果分析 2.1 天祝白牦牛NGB蛋白同源性研究结果显示(见图 1),天祝白牦牛NGB蛋白151个氨基酸残基中,65个氨基酸残基为保守序列,26个氨基酸残基为保守替换,9个氨基酸残基为半保守替换,51个氨基酸残基为未知突变。其中,与NGB蛋白功能相关核心His 64、His 96在进化中为保守序列,Cys 46为未知突变,Cys 55为保守序列;天祝白牦牛两处突变氨基酸,83号位点在进化中为未知突变;97号位点为保守替换。
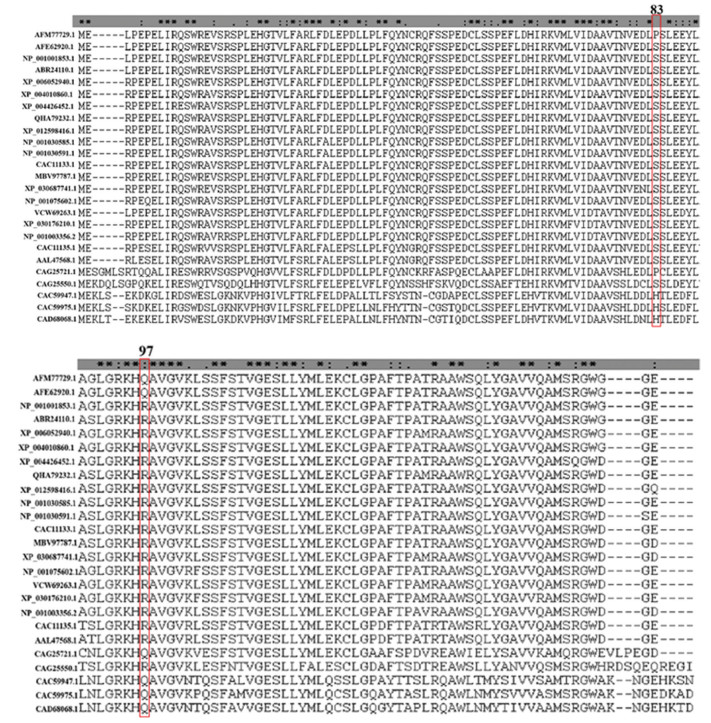
|
图 1 天祝白牦牛与其他物种NGB蛋白氨基酸序列比对 Figure 1 Comparison of amino acid sequences of NGB protein in Tianzhu white yak and other species 注:“*”代表所有比对中相同的残基序列; “: ”代表保守替换; “.”代表半保守替换;AFM77729.1天祝白牦牛,AFE62920.1甘南牦牛,NP_001001853.1黄牛,ABR24110.1藏羚羊,XP_006052940.1印度水牛,XP_004010860.1绵羊,XP_004426452.1白犀牛,QHA79232.1双峰驼,XP_012598416.1鼠狐猴,NP_001030585.1黑猩猩,NP_001030591.1猕猴,CAC11133.1人类,MBV97787.1灰鲸,XP_030687741.1长肢领航鲸,NP_001075602.1穴兔,VCW69263.1貂熊,XP_030176210.1加拿大猞猁,NP_001003356.2家犬,CAC11135.1小鼠,AAL47568.1褐家鼠,CAG25721.1红原鸡,CAG25550.1热带爪蟾,CAC59947.1斑马鱼,CAC59975.1河豚,CAD68068.1虹鳟鱼 Notes: "*" indicates residues that are identical in all the aligned sequences; ": " indicates conserved substitutions; "." indicates semi-conserved substitutions. AFM77729.1 Bos grunniens, AFE62920.1 Bos grunniens, NP_001001853.1 Bos taurus, ABR24110.1 Pantholops hodgsonii, XP_006052940.1 Bubalus bubalis, XP_004010860.1 Ovis aries, XP_004426452.1 Ceratotherium simum simum, QHA79232.1 Camelus bactrianus, XP_012598416.1 Microcebus murinus, NP_001030585.1 Pan troglodytes, NP_001030591.1 Macaca mulatta, CAC11133.1 Homo sapiens, MBV97787.1 Eschrichtius robustus, XP_030687741.1 Globicephala melas, NP_001075602.1 Oryctolagus cuniculus, VCW69263.1 Gulo gulo, XP_030176210.1 Lynx canadensis, NP_001003356.2 Canis lupus familiaris, CAC11135.1 Mus musculus, AAL47568.1 Rattus norvegicus, CAG25721.1 Gallus gallus, CAG25550.1 Xenopus tropicalis, CAC59947.1 Danio rerio, CAC59975.1 Tetraodon nigroviridis, CAD68068.1 Oncorhynchus mykiss |
不同物种NGB蛋白的聚类关系主要分为四大簇,分别为哺乳纲簇、两栖纲簇、鸟纲簇和鱼纲簇,且进化趋于保守(见图 2)。哺乳纲簇中,天祝白牦牛NGB与甘南牦牛、黄牛、印度水牛、绵羊等牛科动物的亲缘关系最近,分支支持率为100%;与白犀牛、穴兔、貂熊、黑猩猩、人类、小鼠等物种的亲缘关系次之,分支支持率大于50%;与鼠狐猴、双峰驼等的亲缘关系较远,分支支持率小于50%。两栖纲簇以热带爪蟾单独聚为一分支,鸟纲簇以红原鸡单独聚为一分支,鱼纲簇以虹鳟鱼、斑马鱼、河豚单独聚为一分支,均与天祝白牦牛NGB亲缘关系较远。
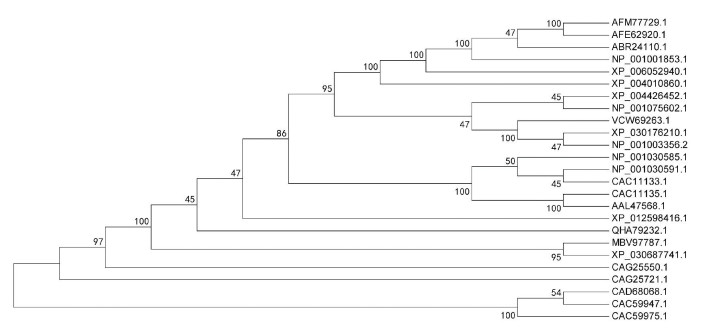
|
图 2 天祝白牦牛与其他物种NGB蛋白系统进化树 Figure 2 Phylogenetic tree of NGB protein in Tianzhu white yak and other species 注:AFM77729.1天祝白牦牛,AFE62920.1甘南牦牛,ABR24110.1藏羚羊,NP_001001853.1黄牛,XP_006052940.1印度水牛,XP_004010860.1绵羊,XP_004426452.1白犀牛,NP_001075602.1穴兔,VCW69263.1貂熊,XP_030176210.1加拿大猞猁,NP_001003356.2家犬,NP_001030585.1黑猩猩,NP_001030591.1猕猴,CAC11133.1人类,CAC11135.1小鼠,AAL47568.1褐家鼠,XP_012598416.1鼠狐猴,QHA79232.1双峰驼,MBV97787.1灰鲸,XP_030687741.1长肢领航鲸,CAG25550.1热带爪蟾,CAG25721.1红原鸡,CAD68068.1虹鳟鱼,CAC59947.1斑马鱼,CAC59975.1河豚. Notes: AFM77729.1 Bos grunniens, AFE62920.1 Bos grunniens, ABR24110.1 Pantholops hodgsonii, NP_001001853.1 Bos taurus, XP_006052940.1 Bubalus bubalis, XP_004010860.1 Ovis aries, XP_004426452.1 Ceratotherium simum simum, NP_001075602.1 Oryctolagus cuniculus, VCW69263.1 Gulo gulo, XP_030176210.1 Lynx canadensis, NP_001003356.2 Canis lupus familiaris, NP_001030585.1 Pan troglodytes, NP_001030591.1 Macaca mulatta, CAC11133.1 Homo sapiens, CAC11135.1 Mus musculus, AAL47568.1 Rattus norvegicus, XP_012598416.1 Microcebus murinus, QHA79232.1 Camelus bactrianus, MBV97787.1 Eschrichtius robustus, XP_030687741.1 Globicephala melas, CAG25550.1 Xenopus tropicalis, CAG25721.1 Gallus gallus, CAD68068.1 Oncorhynchus mykiss, CAC59947.1 Danio rerio, CAC59975.1 Tetraodon nigroviridis. |
分别利用天祝白牦牛与黄牛NGB蛋白在Swiss Model中的不同输出模板,用Spdbv软件对两物种NGB蛋白进行同源建模,得到天祝白牦牛与黄牛NGB蛋白基本结构,如图 3所示。天祝白牦牛与普通黄牛NGB蛋白为血红素蛋白,金属蛋白亚类,卟啉环中心含一个铁原子,该原子在轴两极等距的中纬线上与卟啉环上的四个氮原子以及近端His 96协调配位;在还原状态下,His 64上的一个氮原子与中心铁原子配位,形成双组氨酸六配位结构(见图 3A、3B)。其中,天祝白牦牛近端His 96与中心铁原子距离为2.00 Å,远端His 64与中心铁原子距离为2.10 Å;黄牛近端His 96与中心铁原子距离为1.98 Å,远端His 64与中心铁原子距离为2.17 Å。天祝白牦牛与普通黄牛NGB蛋白三级结构主要是由8个α-螺旋构成的蛋白质多肽,α-螺旋之间的拐弯由1~8个残基构成无规卷曲,其中A,B,E,F,G,H α-螺旋被组装成两层结构,肽链盘绕成球形把血红素分子紧密包裹其中;分子内部几乎全部为疏水性R侧链,分子外表面则包括疏水性和亲水性氨基酸残基,绝大多数亲水性R侧链在分子外表面,使NGB蛋白为可溶于水的蛋白质(见图 3C、3D、3E、3F)。
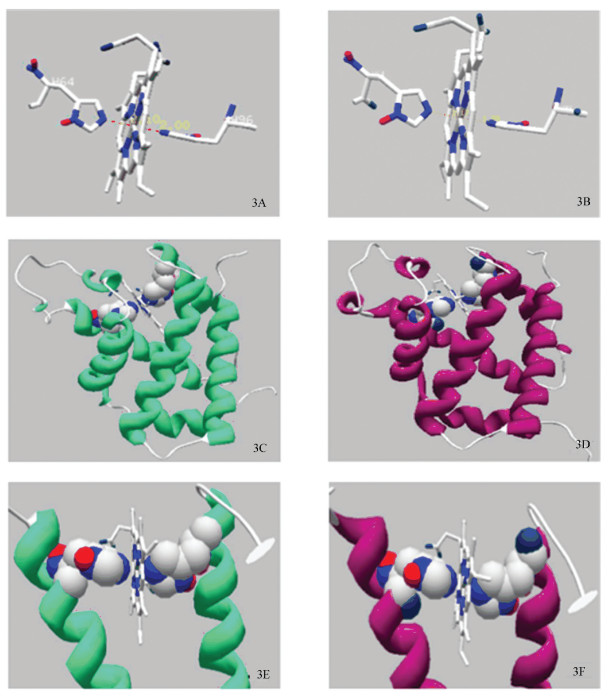
|
图 3 天祝白牦牛与黄牛NGB蛋白基本结构 Figure 3 Basic NGB protein structure of Tianzhu white yak and cattle 注:3A:天祝白牦牛远端His 64和近端His 96与卟啉环中心铁原子立体线性结构;3B:黄牛远端His 64和近端His 96与卟啉环中心铁原子立体线性结构;3C:天祝白牦牛完整NGB蛋白3D结构(绿色);3D:黄牛完整NGB蛋白3D结构(紫色);3E:天祝白牦牛远端His 64和近端His 96与卟啉环中心铁原子3D结构;3F:黄牛远端His 64和近端His 96与卟啉环中心铁原子3D结构. Notes: 3A: Stereoscopic linear structure of the central iron atom and porphyrin ring of distal His 64 and proximal His 96 in Tianzhu white yak; 3B: Stereoscopic linear structure of the central iron atom and porphyrin ring of distal His 64 and proximal His 96 in cattle; 3C: 3D structure of the intact NGB protein in Tianzhu white yak(green). 3D: 3D structure of the intact NGB protein in cattle (purple); 3E: 3D structure of the central iron atom and porphyrin ring of distal His 64 and proximal His 96 in Tianzhu white yak; 3F: 3D structure of the central iron atom and porphyrin ring of distal His 64 and proximal His 96 in cattle. |
NGB蛋白与功能相关核心结构有:血红素“漏斗”结构,腔/隧道结构以及CD环结构。
血红素“漏斗”结构:如图 4A-F所示,NGB蛋白Phe 28,Phe 42、Tyr 44、Lys 67、Val 68构成疏水腔体[13-15],形似“漏斗”,落在血红素缝隙外,通过与血红素丙酸盐的静电作用,参与维持血红素稳定,天祝白牦牛与黄牛NGB蛋白Tyr 44空间取向存在差异(见图 4A、4B、4D、4E),黄牛Tyr 44氧原子指向卟啉环中心。
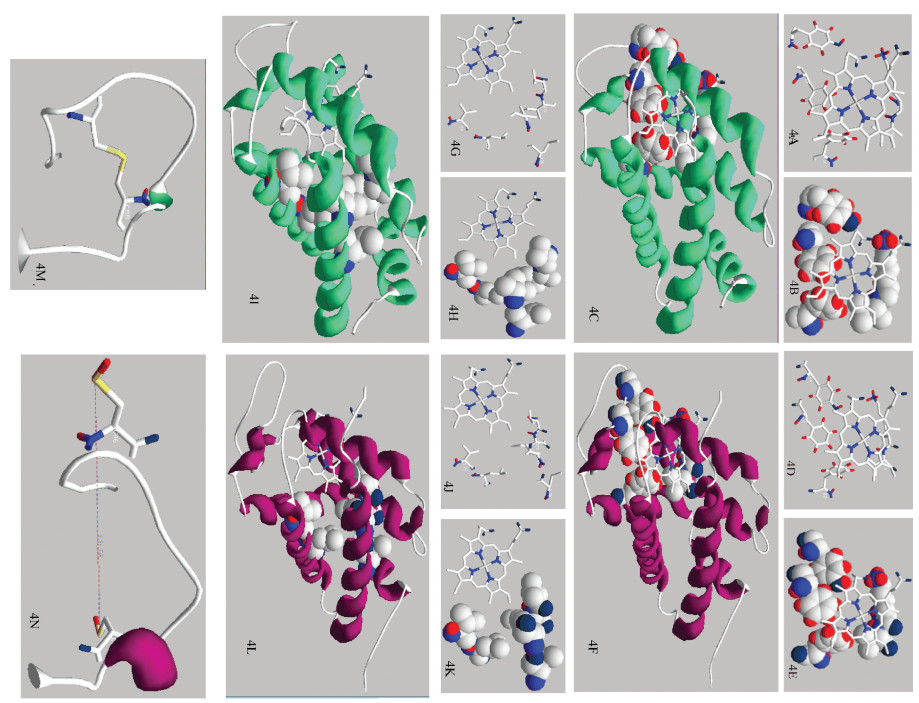
|
图 4 天祝白牦牛与黄牛NGB蛋白与功能相关核心结构 Figure 4 Function-related core structure of NGB protein in Tianzhu white yak and cattle 注:4A、4B:天祝白牦牛NGB血红素“漏斗”特定位点线性及3D结构; 4C:天祝白牦牛NGB血红素“漏斗”完整3D结构;4D、4E:黄牛NGB血红素“漏斗”特定位点线性及3D结构; 4F:黄牛NGB血红素“漏斗”完整3D结构;4G、4H:天祝白牦牛NGB腔/隧道特定位点线性及3D结构; 4I:天祝白牦牛NGB腔/隧道完整3D结构;4J、4K:黄牛NGB腔/隧道特定位点线性及3D结构; 4L:黄牛NGB腔/隧道完整3D结构;4M:天祝白牦牛CD环结构; 4N:黄牛CD环结构. Notes: 4A, 4B: Linear and 3D structure of NGB protein heme "funnel" specific site of Tianzhu white yak; 4C: 3D structure of the intact NGB protein heme "funnel" of Tianzhu white yak; 4D, 4E: Linear and 3D structure of NGB protein heme "funnel" specific site of cattl; 4F: 3D structure of the intact NGB protein heme "funnel" of cattle; 4G, 4H: Linear and 3D structure of NGB protein specific site of cavity/tunnel systems of Tianzhu white yak; 4I: 3D structure of the intact NGB protein cavity/tunnel systems of Tianzhu white yak; 4J, 4K: Linear and 3D structure of NGB protein specific site of cavity/tunnel systems of cattle; 4L: 3D structure of the intact NGB protein cavity/tunnel systems of cattle; 4M: CD loop structure of Tianzhu white yak; 4N: CD loop structure of cattle. |
腔/隧道结构:如图 4G-L所示,NGB腔/隧道结构包括Val 71,Ile 72,Val 109,Leu 113,Leu 136和Val 140[8],该结构在天祝白牦牛和黄牛基本一致。
\CD环结构:天祝白牦牛Cys 46-Cys 55形成二硫键(见图 4M),而黄牛Cys 46、Cys 55未形成二硫键,键距为18.79 Å(见图 4N)。
2.4 天祝白牦牛NGB蛋白突变相关结构对编码NGB蛋白突变位点氨基酸的密码子进行分析,天祝白牦牛NGB蛋白83号位点原Ser密码子分别为UCU、UCC、UCA、UCG,经突变后Pro密码子分别为CCU、CCC、CCA、CCG,主要为1号位U突变为C;97号位点原Arg密码子分别为CGA、CGG,经突变后Gln密码子分别为CAA、CAG,主要为2号位G突变为A。
对NGB蛋白突变位点氨基酸残基的分布特征进行分析,结果列于表 2,3及图 5。由表 2可知,天祝白牦牛和黄牛NGB蛋白突变位点的氨基酸残基中Ser,Pro,Arg,Gln构成α-螺旋和无规卷曲占比不同,在天祝白牦牛中构成α-螺旋分别占:4.6%,2.7%,3.9%,2.7%;在黄牛中分别占:5.3%,2.7%,4.6%,1.9%;在天祝白牦牛中构成无规卷曲分别占:4.6%,3.9%,1.9%,1.3%;在黄牛中分别占:4.6%,3.3%,1.9%,1.3%。
| 表 2 天祝白牦牛与黄牛NGB蛋白突变位点氨基酸残基组成 Table 2 Composition of amino acid residues at the mutation sites of NGB protein in Tianzhu white yak and cattle |
| 表 3 天祝白牦牛与黄牛NGB蛋白Ser、Pro、Arg及Gln分布位点比较 Table 3 Comparison of Ser, Pro, Arg, and Gln sites of NGB protein between Tianzhu white yak and cattle |

|
图 5 天祝白牦牛与黄牛NGB蛋白Ser、Pro、Arg及Gln分布特征比较 Figure 5 Comparison of distribution characteristics of Ser, Pro, Arg and Gln of NGB protein in Tianzhu white yak and cattle 注:5A、5B:天祝白牦牛NGB Ser分布3D结构;5C、5D:天祝白牦牛NGB Pro分布3D结构;5E、5F:黄牛NGB Ser分布3D结构;5G、5H:黄牛NGB Pro分布3D结构;5I、5J:天祝白牦牛NGB Arg分布3D结构;5K、5L:天祝白牦牛NGB Gln分布3D结构;5M、5N:黄牛NGB Arg分布3D结构;5O、5P:黄牛NGB Gln分布3D结构. Notes: 5A, 5B: 3D structure of NGB protein Ser distribution of Tianzhu white yak; 5C, 5D: 3D structure of NGB protein Pro distribution of Tianzhu white yak; 5E, 5F: 3D structure of cattle NGB protein Ser distribution of cattle; 5G, 5H: 3D structure of NGB protein Pro distribution of cattle; 5I, 5J: 3D structure of NGB protein Arg distribution of Tianzhu white yak; 5K, 5L: 3D structure of NGB protein Gln distribution of Tianzhu white yak; 5M, 5N: 3D structure of NGB protein Arg distribution of cattle; 5O, 5P: 3D structure of NGB protein Gln distribution of cattle. |
由表 3和图 5结果可知,天祝白牦牛NGB蛋白Ser多位于α-螺旋的端位,且突变前有4个位点相邻(50、51,57、58,83、84,104、105),醇羟基氧原子指向亲水相(图 5B、5F);NGB蛋白Pro多位于无规卷曲,在第20,52,59,83位点与Ser 19,51,58,84位点相邻,四碳环指向疏水相(图 5D、5H);Arg、Gln多位于α-螺旋的中间位,R侧链氨基端指向亲水相,羧基端参与构成α-螺旋;Arg第10、47位与Gln第11、48位相邻(图 5J、5N、5L、5P)。
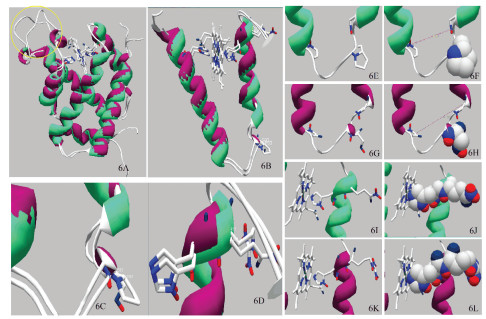
重合天祝白牦牛与黄牛NGB蛋白三级结构,发现重合度较高,但CD环形态(见图 6A,黄色圆圈圈出结构)存在一定差异;天祝白牦牛83号位点为无规卷曲,黄牛83号位点为α-螺旋(见图 6B、6C),97号位点二者均为α-螺旋(见图 6B、6D)。其中,NGB蛋白Ser突变为Pro主要为R侧链由醇羟基突变为四碳环,侧链结构发生改变(见图 6E、6G);突变位于E、F链之间,对使得E、F链之间的间距由11.79Å变为8.21Å(见图 6F、6H);Arg突变为Gln主要为R侧链由氮原子突变为氧原子,且与碳原子通过双键连接,侧链结构未发生改变(见图 6I、6K);97号位点突变基团指向亲水相,羧基端与96号位点相连,对96号位点取向造成影响(见图 6J、6L)。
|
图 6 天祝白牦牛与黄牛NGB蛋白突变位点结构比较 Figure 6 Comparison of mutation site structures of NGB protein in Tianzhu white yak and cattle 注:6A:天祝白牦牛与黄牛NGB重合完整3D结构;6B:天祝白牦牛与黄牛NGB重合E、F链3D结构;6C:天祝白牦牛与黄牛NGB重合83号位点3D结构;6D:天祝白牦牛与黄牛NGB重合97号位点3D结构;6E、6F:天祝白牦牛Pro 83线性及3D结构;6G、6H:黄牛Ser 83线性及3D结构;6I、6J:天祝白牦牛Gln 97线性及3D结构;6K、6L:黄牛Arg 97线性及3D结构. Notes: 6A: Complete 3D structure of NGB superposition in the Tianzhu white yak and cattle; 6B: 3D structure of E and F chains of NGB superposition of Tianzhu white yak and cattle; 6C: 3D structure of NGB superposition of Tianzhu white yak and cattle at 83rdsite; 6D: 3D structure of NGB superposition of Tianzhu white yak and cattle at 97th site; 6E, 6F: Linear and 3D structure of Tianzhu white yak NGB protein Pro 83rd site; 6G, 6H: Linear and 3D structure of cattle of NGB protein Ser 83rd site. 6I, 6J: Linear and 3D structure of Tianzhu white yak of NGB protein Gln at 97th site; 6K, 6L: Linear and 3D structure of cattle NGB protein Arg at 97th site. |
对NGB蛋白的同源序列比较发现,在哺乳动物NGB蛋白中,天祝白牦牛是唯一出现83号位点突变的物种,分析其可能的原因是机体在长期适应高寒低氧环境过程中产生了DNA自发性损伤,编码氨基酸的密码子发生突变,进而导致氨基酸种类由Ser突变为了Pro。比较天祝白牦牛和黄牛NGB蛋白Ser和Pro三级结构的分布特征发现,天祝白牦牛NGB蛋白83号位点Pro偏向于形成无规卷曲(3.9%),而无规卷曲对蛋白质的稳定性和柔韧性均会产生影响[16],因此该突变很可能会对天祝白牦牛NGB蛋白质结构的稳定性和柔韧性带来一定程度的改变。此外,有研究发现Ser具有不带电荷的极性侧链(β羟基),有亲水性,并易被酯化和糖基化;Pro为疏水氨基酸,且与其它标准氨基酸在结构上有很大不同(α亚氨基酸),具有环化的侧链,该侧链对蛋白质的立体结构有很大制约[17],因此,Ser突变形成Pro,对NGB蛋白质溶解度可能会产生一定影响。不过,上述83号突变位点对NGB蛋白产生的一系列影响是否会进一步带来功能上的变化仍需更深入的研究探讨。
本研究结果显示,在还原状态下,天祝白牦牛NGB蛋白可形成双组氨酸六配位结构,Liu等[18]及Hota等[19]研究认为在正常生理条件下,NGB约占细胞总浓度的30%,在低氧条件下,许多神经保护性低氧诱导型基因(如低氧诱导因子-1α,血管内皮生长因子,血红素加氧酶-1和促红细胞生成素)上调以协助促进细胞存活,这使得NGB浓度迅速上升到原浓度的80%,且这种上升发生在数秒之间,说明NGB蛋白在还原状态下大量稳定存在有利于机体适应高原低氧环境,而83号位点突变造成了E、F链距离变窄,致使核心His 64与卟啉环中心距离变短(由2.17Å变为2.10Å),使还原状态下天祝白牦牛NGB蛋白结构更为稳定,这可能是NGB蛋白在牦牛适应高原低氧环境中发挥作用的重要结构基础。有研究报道显示,NGB蛋白与外源性配体如O2、CO、NO等发生结合时,首先要打破铁和远端His 64之间的纽带,而后外源性配体才有可能与NGB结合,外源性配体与内源性远端His 64之间的竞争导致了配体结合动力学的复杂模式[20],因此His 64位点与卟啉环中心距离的改变极有可能对血红素外源配体的结合产生一定的影响。由此,83号位点突变很有可能是天祝白牦牛适应高寒低氧环境所必须的,后续实验可继续针对天祝白牦牛该突变位点与NGB蛋白配体结合能力之间的联系开展相应研究工作。
与83号位点相比较,天祝白牦牛NGB蛋白97号位点的突变是保守替换,虽然导致了氨基酸种类发生改变,但对NGB蛋白结构并未产生影响,不过研究显示Arg等电点为10.76,而Gln等电点为5.65[17],差异较大,提示该位点突变也会对NGB蛋白的溶解度产生一定影响。此外,97号位点突变使核心His 96与卟啉环中心距离由1.98 Å变为2.00 Å,说明该突变对96号位点的向心取向会造成一定改变,由于His 96是整个NGB蛋白最核心的位点,决定着NGB蛋白的活性[21],因此向心取向的变化可能会对NGB蛋白功能的发挥造成影响,究其具体影响范围和机制尚有待进一步试验验证。总体而言,97号位点突变虽然对蛋白质基本结构不造成影响,却改变了蛋白核心位点的向心取向,同时对蛋白质溶解度也造成了一定影响,此外,与天祝白牦牛亲缘关系最近的甘南牦牛也存在97号位点的突变现象,对该位点突变的深入探究同样具有十分重要的意义。
相较于黄牛,天祝白牦牛NGB蛋白两处氨基酸突变对其血红素“漏斗”结构及腔/隧道结构均未造成影响,说明两结构在进化中趋于保守,但CD环结构存在一定差异。研究表明,NGB蛋白CD环上的Cys 46、Cys 55因其R基团具有-SH,因此可在空间形成二硫键,Cys 46-Cys 55键的形成可降低血红素-Fe原子的第六配位键的强度,促进配体结合;相反,Cys 46-Cys 55键的裂解可稳定血红素-Fe原子的六配位键并损害金属中心的反应性[22-25],天祝白牦牛Cys 46-Cys 55键为结合状态,而黄牛为断裂状态(键距:18.79Å),说明天祝白牦牛NGB蛋白配体结合能力更强,鉴于目前有大量NGB蛋白结构学研究集中在对其CD环结构的研究上[9, 10, 23, 24],也进一步印证了其重要性,天祝白牦牛相较于黄牛CD环结构的差异是否与两处突变位点有关联仍有待更进一步的分析验证。
| [1] |
BURMESTER T, WEICH B, REINHARDT S, et al. A vertebrate globin expressed in the brain[J]. Nature, 2000, 407(6803): 520-523. DOI:10.1038/35035093 (  0) 0) |
| [2] |
ZHANG C G, WANG C L, DENG M Y, et al. Full-length cDNA cloning of human neuroglobin and tissue expression of rat neuroglobin[J]. Biochemical & Biophysical Research Communications, 2002, 290(5): 1410-1419. DOI:10.1006/bbrc.2002.6360 (  0) 0) |
| [3] |
张黎. 外膜蛋白功能性结构研究[D]. 长春: 吉林大学, 2017. ZHANG Li. Study on the functional structure of outer membrane protein[D]. Changchun: Jilin University, 2017. (  0) 0) |
| [4] |
GAO Y, YIN H, ZHANG Y F, et al. Dexmedetomidine protects hippocampal neurons against hypoxia/reoxygenation-induced apoptosis through activation HIF-1α/p53 signaling[J]. Life Sciences, 2019, 232: 116611. DOI:10.1016/j.lfs.2019.116611 (  0) 0) |
| [5] |
ZHANG Y F, YANG F, GAO Y, et al. Neuroglobin protects offspring rats from neuronal damage induced by sevoflurane exposure to pregnant rats by inhibiting endogenous apoptosis[J]. International Journal of Developmental Neuroscience, 2019, 76(1): 17-24. DOI:10.1016/j.ijdevneu.2019.06.001 (  0) 0) |
| [6] |
BAEZ-JURADO E, GUIO-VEGA G, HIDALGO-LANUSSA O, et al. Mitochondrial neuroglobin is necessary for protection induced by conditioned medium from human adipose-derived mesenchymal stem cells in astrocytic cells subjected to scratch and metabolic injury[J]. Molecular Neurobiology, 2019, 56(7): 5167-5187. DOI:10.1007/s12035-018-1442-9 (  0) 0) |
| [7] |
FIOCCHETTI M, CIPOLLETTI M, ASCENZI P, et al. Dissecting the 17β-estradiol pathways necessary for neuroglobin anti-apoptotic activity in breast cancer[J]. Journal of Cellular Physiology, 2018, 233(7): 5087-5103. DOI:10.1002/jcp.26378 (  0) 0) |
| [8] |
GUIMARAES B G, HAMDAN E D, LECHAUVE C, et al. The crystal structure of wild-type human brain neuroglobin reveals flexibility of the disulfide bond that regulates oxygen affinity[J]. Acta Crystallographica. Section D, Biological Crystallography, 2014, 70(1): 1005-1014. DOI:10.1107/S1399004714000078 (  0) 0) |
| [9] |
LEI R, BUTCHER D, BERNAD S, et al. Mutation of residues in CD loop and distal pocket impact protein stability of human neuroglobin[J]. Biophysical Journal, 2019, 116(3): 1256-1265. DOI:10.1016/j.bpj.2018.11.2545 (  0) 0) |
| [10] |
EXERTIER C, MILAZZO L, FREDA I, et al. Proximal and distal control for ligand binding in neuroglobin: Role of the CD loop and evidence for His64 gating[J]. Scientific Reports, 2019, 9(1): 5326. DOI:10.1038/s41598-019-41780-3 (  0) 0) |
| [11] |
郭琳, 钟金城, 柴志欣, 等. 西藏牦牛NGB基因克隆及生物信息学分析[J]. 江苏农业科学, 2015, 43(11): 26-30. GUO Lin, ZHONG Jincheng, CHAI Zhixin, et al. NGB gene cloning and bioinformatics analysis of Tibetan yak[J]. Jiangsu Agricultural Sciences, 2015, 43(11): 26-30. (  0) 0) |
| [12] |
李盛杰, 杜晓华, 罗玉柱, 等. 天祝白牦牛NGB基因的克隆及生物信息学分析[J]. 畜牧兽医学报, 2013, 44(3): 395-398. LI Shengjie, DU Xiaohua, LUO Yuzhu, et al. Tianzhu white yak NGB gene cloning and bioinformatics analysis[J]. Journal of Animal Science and Veterinary Medicine, 2013, 44(3): 395-398. (  0) 0) |
| [13] |
ARCOVITO A, MOSCHETTI T, D'ANGELO P, et al. An X-ray diffraction and X-ray absorption spectroscopy joint study of neuroglobin[J]. Archives of Biochemistry & Biophysics, 2008, 475(1): 7-13. DOI:10.1016/j.abb.2008.03.026 (  0) 0) |
| [14] |
AVELLA G, ARDICCIONI C, SCAGLIONE A, et al. Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism[J]. Acta Crystallographica Section D, 2014, 70(6): 1640-1648. DOI:10.1107/S1399004714007032 (  0) 0) |
| [15] |
VALLONE B, NIENHAUS K, BRUNORI M, et al. The structure of murine neuroglobin: Novel pathways for ligand migration and binding[J]. Proteins, 2004, 56(1): 85-92. DOI:10.1002/prot.20113 (  0) 0) |
| [16] |
王镜岩, 朱圣庚, 徐长法. 生物化学[M]. 北京: 高等教育出版社, 2002: 152-177. WANG Jingyan, ZHU Shenggeng, XU Changfa. Biochemistry[M]. Beijing: Higher Education Press, 2002: 152-177. (  0) 0) |
| [17] |
邹思湘. 动物生物化学[M]. 北京: 中国农业出版社, 2015: 35-50. ZOU Sixiang. Animal biochemistry[M]. Beijing: China Agricultural Press, 2015: 35-50. (  0) 0) |
| [18] |
LIU A, BRITTAIN T. A futile redox cycle involving neuroglobin observed at physiological temperature[J]. International Journal of Molecular Sciences, 2015, 16(8): 20082-20094. DOI:10.3390/ijms160820082 (  0) 0) |
| [19] |
HOTA K B, HOTA S K, SRIVASTAVA R B, et al. Neuroglobin regulates hypoxic response of neuronal cells through Hif-1α-and Nrf2-mediated mechanism[J]. Journal of Cerebral Blood Flow & Metabolism, 2012, 32(6): 1046-1060. DOI:10.1038/jcbfm.2012.21 (  0) 0) |
| [20] |
李直, 李依芃, 邓锦波. 脑红蛋白对神经保护作用的研究[J]. 河南大学学报(医学版), 2016, 35(3): 225-228. LI Zhi, LI Yipeng, DENG Jinbo. Study of neuroprotective effect of cerebrin[J]. Journal of Henan University (Medical Sciences), 2016, 35(3): 225-228. (  0) 0) |
| [21] |
ASCENZI P, MASI A, LEBOFFE L, et al. Neuroglobin: From structure to function in health and disease[J]. Molecular Aspects of Medicine, 2016, 52(1): 1-48. DOI:10.1016/j.mam.2016.10.004 (  0) 0) |
| [22] |
PESCE A, DEWILDE S, NARDINI M, et al. Human brain neuroglobin structure reveals a distinct mode of controlling oxygen affinity[J]. Structure, 2003, 11(9): 1087-1095. DOI:10.1016/s0969-2126(03)00166-7 (  0) 0) |
| [23] |
BURMESTER T, HANKELN T. Neuroglobin: A respiratory protein of the nervous system[J]. News Physiologia Science, 2004, 19(1): 110-113. DOI:10.1152/nips.01513.2003 (  0) 0) |
| [24] |
LABERGE M, YONETANI T. Common dynamics of globin family proteins[J]. IUBMB Life, 2007, 59(8/9): 528-534. DOI:10.1080/15216540701222914 (  0) 0) |
| [25] |
HAMDANE D, KIGER L, DEWILDE S, et al. The redox state of the cell regulates the ligand binding affinity of human neuroglobin and cytoglobin[J]. Journal of Biological Chemistry, 2003, 278(51): 51713-51721. DOI:10.1074/jbc.M309396200 (  0) 0) |
 2021, Vol. 19
2021, Vol. 19


